Team:ETH Zurich/Project/Approach
From 2008.igem.org
(→Our Approach) |
(→Our Approach) |
||
| Line 22: | Line 22: | ||
We start by inoculating our continuous culture with a wild type strain with a knock out that makes it auxotroph for nucleotides. These nucleotides will be the limiting substrate in the chemostat and the concentration in our feed will be the parameter we can tune to set the selective pressure. See the [[Team:ETH Zurich/Wetlab/Chemostat Selection| chemostat selection]] section for a detailed description of the selection process. | We start by inoculating our continuous culture with a wild type strain with a knock out that makes it auxotroph for nucleotides. These nucleotides will be the limiting substrate in the chemostat and the concentration in our feed will be the parameter we can tune to set the selective pressure. See the [[Team:ETH Zurich/Wetlab/Chemostat Selection| chemostat selection]] section for a detailed description of the selection process. | ||
| + | |||
| + | The iteration of the following steps gradually reduces the size of the chromosome: | ||
| + | |||
| + | '''1) Excision of genomic sequences by restriction enzymes and ligases.''' We have developed a [[Team:ETH Zurich/Modeling/Switch Circuit|genetic circuit]] that allows us to generate a controlled pulse to control the expression of restriction enzymes and ligases. | ||
Revision as of 01:15, 30 October 2008
|
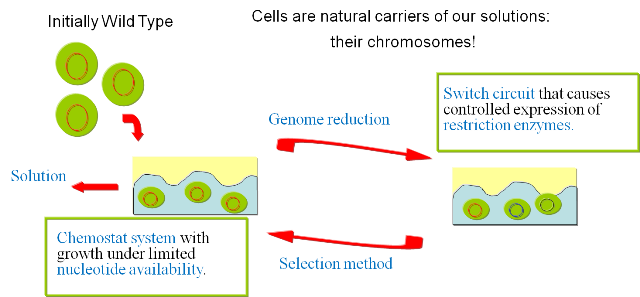
Our ApproachWe are following an approach based on random genome deletions and a designed selection mechanism to reach our goal. As mentioned in the Motivation section, we postulate that a random disruption of genes combined with an evolutionary approach will allow us to dramatically reduce the genome size of an organism. In order to prove the viability of the concept, we have developed a series of models and of biological experiments that are introduced in this section. For details on the results of the single steps we refer to the specific sections in the Wetlab and the Modelingsections. Figure 1 gives an overview of the developed system and illustrates the final experimental setup.
The iteration of the following steps gradually reduces the size of the chromosome: 1) Excision of genomic sequences by restriction enzymes and ligases. We have developed a genetic circuit that allows us to generate a controlled pulse to control the expression of restriction enzymes and ligases.
See the chemostat model section for a detailed description of the selection process. a) .. excision of genomic sequences by restriction enzymes can be performed within the living cell; b) .. fragmented genomic DNA can be ligated inside the living cell by simultaneous expression of T4 ligase; c) .. pulse generators that produce pulses of protein expression of variable length terminated by induction with a second compound can be devised both on the transcriptional as well as on the translational level; d) .. cells with optimized growth and reproduction can be selected by competition inside chemostat cultures. Random walk towards a minimal genome |
 "
"