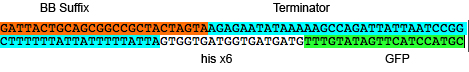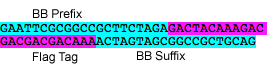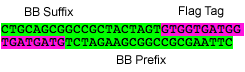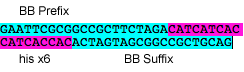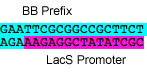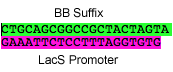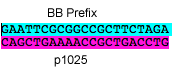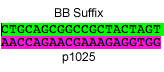Team:MIT/sequences
From 2008.igem.org
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
Primers
- All BioBrick parts are Phillips-Silver to enable fusion peptide construction
P1: Forward Primer for Promoter
- foward sequence: CCG CTT CTA GAG TAA TAC GAC TCA CTA TAG GGA ATA CAA GCT ACT TGT TCT TTT TGC ATA CTA GAG ATT AAA GAG GAG AAA TAC TAG ATG CAG AAG AAA AAA TCC GC
- length = 107 bp
- GC content = 37.4 %
- melting temp = 68.9 ºC
- concentration:
- original:
- dilute: 5uM
P2: Reverse Primer for Peptide
- reverse sequence: GTT CTT CTC CTT TAC GCA TGC TGC CCC CGC CGC TGC CGC C
- length = 40 bp
- GC content = 67.5 %
- melting temp = 74.7 ºC
- concentration:
- original:
- dilute: 5uM
P3: Forward Primer for GFP
- forward sequence: GGC GGC AGC GGC GGG GGC AGC ATG CGT AAA GGA GAA GAA C
- length = 40 bp
- GC content = 67.5 %
- melting temp = 74.7 ºC
- concentration:
- original:
- dilute: 5μM
P4: Reverse Primer for GFP
- sequence: CTG CAG CGG CCG CTA CTA GTA AGA GAA TAT AAA AAG CCA GAT TAT TAA TCC GGC TTT TTT ATT ATT TTT ATT AGT GGT GAT GGT GAT GAT GTT TGT ATA GTT CAT CCA TGC
- length = 111 bp
- GC content % = 36.0 %
- melting temp = 69.6 ºC
- concentration =
P5: Reverse Primer for GFP without Terminator
- sequence: CTG CAG CGG CCG CTA CTA GTA TTA TTA GTG GTG ATG GTG ATG ATG TTT GTA TAG TTC ATC CAT GC
- length = 65bp
- GC content = 44.6 %
- melting temp = 68.5 ºC
- concentration:
P6: Forward Primer for BioBrick FLAG tag
- sequence: GAATTCGCGGCCGCTTCTAGAGACTACAAAGACGACGACGACAAAACTAGTAGCGGCCGCTGCAG
- length = 65bp
- GC content = 55.4%
- melting temp = 72.1 ºC
- concentration =
P7: Reverse Primer for BioBrick FLAG tag
- sequence: CTGCAGCGGCCGCTACTAGTTTTGTCGTCGTCGTCTTTGTAGTCTCTAGAAGCGGCCGCGAATTC
- length = 65bp
- GC content = 55.4%
- melting temp = 72.1 ºC
- concentration =
P8: Forward Primer for BioBrick HA tag
- sequence: GAATTCGCGGCCGCTTCTAGATACCCGTACGACGTTCCGGACTACGCTACTAGTAGCGGCCGCTGCAG
- length = 68bp
- GC content = 60.3%
- melting temp = 73.4 ºC
- concentration =
P9: Reverse Primer for BioBrick HA tag
- sequence:CTGCAGCGGCCGCTACTAGTAGCGTAGTCCGGAACGTCGTACGGGTATCTAGAAGCGGCCGCGAATTC
- length = 68bp
- GC content = 60.3%
- melting temp = 73.4 ºC
- concentration =
P10: Forward Primer for BioBrick His tag
- sequence: GAATTCGCGGCCGCTTCTAGACATCATCACCATCACCACACTAGTAGCGGCCGCTGCAG
- length = 59bp
- GC content = 57.6%
- melting temp = 72.7 ºC
- concentration =
P11: Reverse Primer for BioBrick His tag
- sequence: CTGCAGCGGCCGCTACTAGTGTGGTGATGGTGATGATGTCTAGAAGCGGCCGCGAATTC
- length = 59bp
- GC content = 57.6%
- melting temp = 72.7 ºC
- concentration =
P12: Forward Primer for BioBrick TEV protease cut site
- sequence: GAATTCGCGGCCGCTTCTAGAGAAAACCTGTACTTCCAGGGTACTAGTAGCGGCCGCTGCAG
- length = 62bp
- GC content = 56.5%
- melting temp = 72.2 ºC
- concentration =
P13: Reverse Primer for BioBrick TEV protease cut site
- sequence: CTGCAGCGGCCGCTACTAGTACCCTGGAAGTACAGGTTTTCTCTAGAAGCGGCCGCGAATTC
- length = 62bp
- GC content = 56.5%
- melting temp = 72.2 ºC
- concentration =
P14: Forward Primer for BioBrick pRT B l.bulgaricus signal sequence
- sequence = GAATTCGCGGCCGCTTCTAGAATGCAGAAGAAAAAATCCGC
- length = 41bp
- GC content = 48.8%
- melting temp = 67.1 ºC
- concentration =
P15: Reverse Primer for BioBrick prt B l.bulgaricus signal sequence
- sequence = CTGCAGCGGCCGCTACTAGTGGTAACCGGAGCCGTTTCTTG
- length = 41bp
- GC content = 61%
- melting temp = 71.3 ºC
- concentration =
P16: Forward Primer for BioBrick L. Bulgaricus lacS promoter
- sequence = GAATTCGCGGCCGCTTCTAGAGAAGAGGCTATATCGC
bold = biobrick prefix
- length = 37 bp
- GC content = 54.1%
- melting temp = 66.6 ºC
P17: Reverse Primer for BioBrick L. Bulgaricus lacS promoter
- sequence = CTGCAGCGGCCGCTACTAGTAGAAATTCTCCTTTAGGTGTG
bold = biobrick suffix
- length = 41 bp
- GC content = 51.2%
- melting temp = 66.7 ºC
P18: Forward Primer for BioBrick p1025
- sequence = GAATTCGCGGCCGCTTCTAGACAGCTGAAAACCGCTGACCTG
- length = 42bp
- GC content = 57.1%
- melting temp = 70.2ºC
P19: Reverse Primer for BioBrick p1025
- sequence = CTGCAGCGGCCGCTACTAGTAACCAGAACGAAAGAGGTGG
- length = 40 bp
- GC content = 57.5%
- melting temp = 69.2ºC
P20: Forward Primer for BioBrick GFP
- sequence = GAATTCGCGGCCGCTTCTAGACGTAAAGGAGAAGAACTTTTC
- length = 42 bp
- GC content = 47.6%
- melting temp = 65.9ºC
P21: Reverse Primer for BIoBrick GFP
- sequence = CTGCAGCGGCCGCTACTAGTTTTGTATAGTTCATCCATGC
- length = 40bp
- GC content = 50%
- melting temp = 66.5 ºC
P22: Forward Primer for BBa_I13521
- Will basically remove the promoter from this RFP device
- sequence = gaattcgcggccgcttctagagtactagagaaagaggagaa
- length = 41 bp
- GC content = 48.8%
- melting temp = 65.9 C
P23: Reverse Primer for BBa_I13521
- sequence = ctgcagcggccgctactagtatataaacgcagaaaggccca
- length = 41 bp
- GC content = 53.7%
- melting temp = 68.9 C
Plasmids
Include sequence, BioBrick #, functional features, digestion map.
PCR Products
1-1 GFP
GFP with linker @ 3' and [His, Terminator, Suffix] @ 5'
1-2 GFP
GFP with linker @ 3' and [His, Suffix] @ 5'
2-1 p1025
p1025 construct with [prefix, promoter, rbs] @ 3' and linker @ 5'
2-2 signal
L. bulgaricus signal peptide with Silver modified BioBricks prefix and suffix
2-3 Flag
Flag tag with Silver modified BioBricks prefix and suffix
2-4 HA
HA tag with Silver modified BioBricks prefix and suffix
2-5 His
His tag with Silver modified BioBricks prefix and suffix
2-6 Tev
Tev cleavage site with Silver modified BioBricks prefix and suffix
Other
TEV Protease cut site
- Amino Acid Sequence : Glu-Asn-Leu-Tyr-Phe-Gln-Gly (Cuts between Gln-Gly)
- Base Pair Sequence: GAAAACCTGTACTTCCAGGGT
DNA sequence to synthesize - epitope/p1025/linker (6/17/08)
- The signal peptide sequence (141 bp) can be found at [http://www.ncbi.nlm.nih.gov/sites/entrez?dispmax=1&db=nucleotide&doptcmdl=graph&term=104773257 the NCBI genome database]
- 6/17, lengthened signal peptide so that it will cleave properly
- silent mutations in linkers to reduce repetition and to ensure specificity of primers (ex. GGC/GGA/GGG instead of GGT coding for Gly)
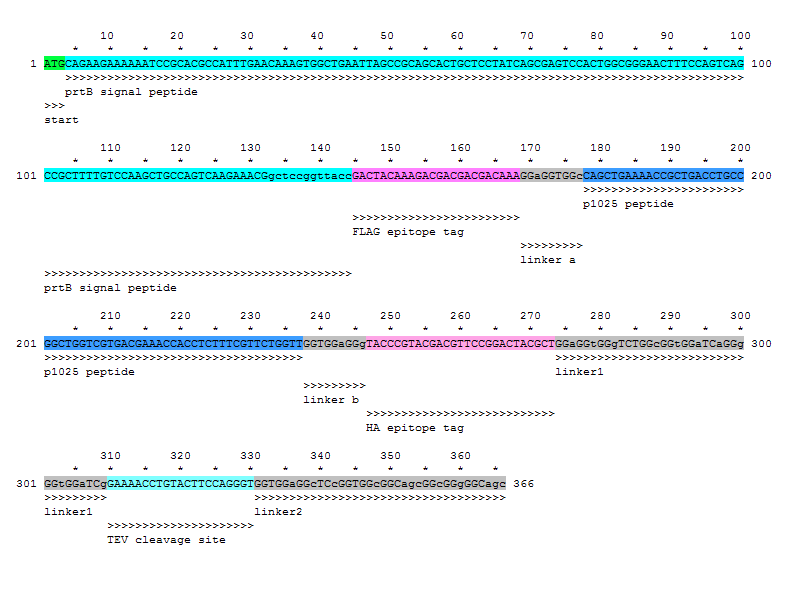
sequence: ATGCAGAAGAAAAAATCCGCACGCCATTTGAACAAAGTGGCTGAATTAGCCGCAGCACTGCTCC TATCAGCGAGTCCACTGGCGGGAACTTTCCAGTCAGCCGCTTTTGTCCAAGCTGCCAGTCAAGA AACGgctccggttaccGACTACAAAGACGACGACGACAAAGGaGGTGGcCAGCTGAAAACCGCT GACCTGCCGGCTGGTCGTGACGAAACCACCTCTTTCGTTCTGGTTGGTGGaGGgTACCCGTACG ACGTTCCGGACTACGCTGGaGGtGGgTCTGGcGGtGGaTCaGGgGGtGGaTCgGAAAACCTGTA CTTCCAGGGTGGTGGaGGcTCcGGTGGcGGCagcGGcGGgGGCagc
L. bulgaricus lacS promoter to synthesize (7/11/08)
sequence:
GAATTCGCGGCCGCTTCTAGAGAAGAGGCTATATCGCCATCATTAGCAGCTTAATTGA ATATTTACTGGCTAAACTATTGAGTTTTCAAGGCTTCATAGTTCTTTTTGGTGTGGGA AGTTTAAATTACTAAAAATATTTTAGTAAAACATCTTGGTTTATTTAGTAAACAAGTC TATACTGTAATTATAAACAAGTTAACACACCTAAAGGAGAATTTCTACTAGTAGCGGC CGCTGCAG
Whole Sequence
CCGCTTCTAGAGTAATACGACTCACTATAGGGAATACAAG CTACTTGTTCTTTTTGCATACTAGAGATTAAAGAGGAGAA ATACTAGATGCAGAAGAAAAAATCCGCACGCCATTTGAAC AAAGTGGCTGAATTAGCCGCAGCACTGCTCCTATCAGCGA GTCCACTGGCGGGAACTTTCCAGTCAGCCGCTTTTGTCCA AGCTGCCAGTCAAGAAACGGCTCCGGTTACCGACTACAAA GACGACGACGACAAAGGaGGTGGcCAGCTGAAAACCGCTG ACCTGCCGGCTGGTCGTGACGAAACCACCTCTTTCGTTCT GGTTGGTGGaGGgTACCCGTACGACGTTCCGGACTACGCT GGaGGtGGgTCTGGcGGtGGaTCaGGgGGtGGaTCgGAAA ACCTGTACTTCCAGGGTGGTGGaGGcTCcGGTGGcGGCag cGGcGGgGGCagcATGCGTAAAGGAGAAGAACTTTTCACT GGAGTTGTCCCAATTCTTGTTGAATTAGATGGTGATGTTA ATGGGCACAAATTTTCTGTCAGTGGAGAGGGTGAAGGTGA TGCAACATACGGAAAACTTACCCTTAAATTTATTTGCACT ACTGGAAAACTACCTGTTCCATGGCCAACACTTGTCACTA CTTTCGGTTATGGTGTTCAATGCTTTGCGAGATACCCAGA TCATATGAAACAGCATGACTTTTTCAAGAGTGCCATGCCC GAAGGTTATGTACAGGAAAGAACTATATTTTTCAAAGATG ACGGGAACTACAAGACACGTGCTGAAGTCAAGTTTGAAGG TGATACCCTTGTTAATAGAATCGAGTTAAAAGGTATTGAT TTTAAAGAAGATGGAAACATTCTTGGACACAAATTGGAAT ACAACTATAACTCACACAATGTATACATCATGGCAGACAA ACAAAAGAATGGAATCAAAGTTAACTTCAAAATTAGACAC AACATTGAAGATGGAAGCGTTCAACTAGCAGACCATTATC AACAAAATACTCCAATTGGCGATGGCCCTGTCCTTTTACC AGACAACCATTACCTGTCCACACAATCTGCCCTTTCGAAA GATCCCAACGAAAAGAGAGACCACATGGTCCTTCTTGAGT TTGTAACAGCTGCTGGGATTACACATGGCATGGATGAACT ATACAAACATCATCACCATCACCACTAATAAAAATAATAA AAAAGCCGGATTAATAATCTGGCTTTTTATATTCTCTTAC TAGTAGCGGCCGCTGCAGGATTA
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
 "
"