Team:Heidelberg/Human Practice/Phips the Phage/General Backround
From 2008.igem.org

...I am glad you decided to learn something from me. You won’t regret it!
Synthetic Biology
Synthetic Biology is a young field in science, which combines classic gene technology with an engineering approach. Similar to the construction of an airplane, Synthetic Biology uses simple gene building blocks for the construction of new complex systems with distinct functions.
So doing Synthetic Biology is like playing with Lego: You have multiple parts, but instead of being made of plastic, they consist of DNA. DNA is a long molecule which can be imagined as being a thin and long peace of paper on which in Morse is printed a construction plan or and advise. DNA can contain the information to build up molecules like proteins, or it can have a regulatory function. As DNA is only an information-housing building it is totally useless if there is nothing reading out the information. And that is why you have to transfer DNA into cells if you want to examine the information a DNA molecule contains.
What synthetic biology does is combining DNA-molecules housing different, well-known information to engineer systems carrying out a certain function. By combining for example a regulatory part that initiates the reading of a gene very efficiently together with a part that contains the information to make a green fluorescent protein, you can built a more complex device, which is able to synthesize GFP at a high rate. If you transfer this device into a bacterial cell, you get green glowing cells, which is quite cool. By transferring different fluorescent-coding devices into different cells you could i.e. built a different colored mirror ball, which is really fancy. But since mirror balls already exist, synthetic biology tries to engineer systems carrying out more usable functions, like the production of biofuels or energy, the degradation of polluting substances or systems able to carry out a medical function. These gene building blocks are collected in databases; so many people can profit from them and expand the range of available parts by their own. Such a database can be imagined as the Lego-Box of the modern synthetic biologist, containing DNA-molecules (the so-called biobricks), used to built up the different systems. The general aim of Synthetic biology is to make a system which provides chassis bacteria that only contain the genes they need to survive under laboratory conditions. They won’t have any additional functions that could need additional energy and resources. Together with this chassis, you would need the set of genes you want to use. They all will be in a form of standardized BioBrick. If you want your bacterium to have any function besides surviving, like producing an enzyme or a drug, you can put this part into it, grow it and harvest you protein of interest. By this approach, the manufacturing of recombinant proteins can be made much more efficient (and therefore cheaper) than with classic biotechnology.
iGEM
"Can simple biological systems be built from standard, interchangeable parts and operated in living cells? Or is biology just too complicated to be engineered in this way?" - Randy Rettberg, director of the iGEM competition
The international competition for Genetically Engineered Machines is one of the biggest student competitions in Synthetic Biology. The general idea of the founders in 2004 was to use the student laboratories at the universities, which normally are empty through the summer months, for a meaningful purpose. This purpose is to set up and carry out a project in the field of Synthetic Biology by undergraduates, who are advised by graduate student and post docs. The competition has rapidly expanded: Starting with 5 teams in the year 2004 to 82 teams in 2008. The aim of the competition is not only to gain experience in the lab and with scientific research, but also to be a pioneer in synthetic biology. This is why the general setup of the competition includes tools and concepts for ideal synthetic biological work: All the parts that are made by the students have to be brought into a standardized form and will be put into a database called registry. The participating teams are invited to use the existing parts in the registry – more than 1000 so far – for their project and also contribute new ones. Thereby the registry expands over the years and the existing parts become tested and improved. Every team participating in the competition got the whole registry before the iGEM-competition started. The registry is just a simple folder where the different DNA-molecules (biobricks) are printed on pieces of paper and are ready to be transferred into organisms. If you want to learn more about the Lego-Box of the synthetic biologists just follow me…
Basics about genes and proteins
Is it impossible to take the gene of an animal, say the one that encodes for the proteins to produces insulin, put it into a bacteria cell and let this cell produce insulin itself? No, it’s not. This is what biotechnologists do every day. They take genes they need for their research or for the production of certain proteins and put them into bacteria cells, because these cells grow very fast, can be cultivated easily and can produce the protein of interest which the researcher can then harvest. But how is this done? Well, to understand this technique, we have to recapitulate the basics of DNA production and information encoding by DNA. As you may well know, DNA (deoxyribonucleic acid) encodes the genetic information of organisms- of all organisms known in the world, by the way. In particular, DNA encodes the information to make proteins. Proteins are molecules which carry out all the major functions of the organism and build its major structures, so they are the building blocks of any organism. (Of course carbon hydrates and lipids are also important, but everything an organism does with them, like uptake, metabolism, assembly, reorganization, degradation, is carried out by proteins.)
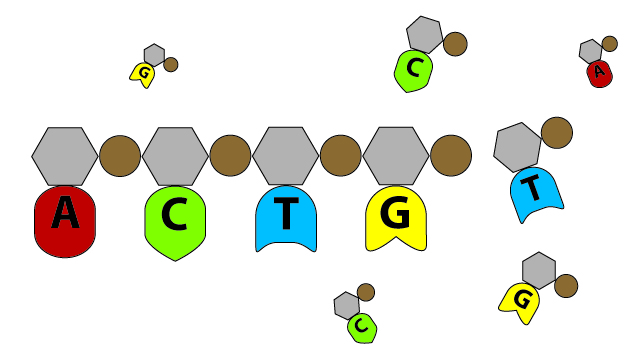
The way the DNA encodes proteins is simple and genius: The DNA consists of 4 different building blocks, the so-called nucleotides. Each nucleotide consists of a sugar (desoxyribose), a phosphate residue and, what is most interesting, a variable nucleotide base. The nucleotide base can be adenine (A), guanine (G), cytosine (C) or thymine (T), so that we get four different possibilities of building up our nucleotide. The important thing for information encoding is the DNA sequence, which is formed by the assembly of different nucleotides into a (linear) DNA strand.
This sequence contains the information for building up a protein. That's only possible, because proteins themselves consist of different, basic building blocks, called amino acids. But instead of just being built of 4 building blocks proteins contain 20 different amino acids. As you might guess, that is a mathematical problem: How can 4 different nucleotides our DNA contains, store information for proteins containing 20 different amino acids? Well the answer is simple: The DNA is organized in subgroups of 3 nucleotides, called codons. By that the DNA houses 64 (4 to the power of 3) different codons, each representing one amino acid.
Now you might again wonder: If I only need 20 codons for encoding 20 amino acids, what role play the other 44 remaining ones? Well, again the answer is simple: Most amino acids are not encoded by one, but by two or even four codons. So there are many possible DNA sequences encoding the same amino acid sequence and by that the same protein. This property of the DNA code is called redundancy. A DNA strand of 300 nucleotides can thus contain information for building a protein of 100 amino acids.
Another interesting thing about DNA is the fact, that it always consists of 2 strands containing the same information. These two strands look alike very much. The only difference is, that at each position the main strand (coding strand) contains the right nucleotides (right in sense of amino acid coding) whereas the other (complementary) strand contains a nucleotide which is able to bind to the nucleotide of the coding strand at a certain position. Concrete: if the coding strand contains an A, the complementary strand contains a T. And if the coding strand contains a G, the complementary strand contains a C. So A and T, C and G match like a key and its lock.
By that, the information on DNA is quite secure, because it is stored two times: On the coding and on the complementary stand. And if one strand gets damaged, the other one still contains the whole information needed to build up a certain protein. So there is always a safety copy in our DNA.
As the complementary strand can bin the coding strand perfectly at each nucleotide position, they form a double strand- the typical DNA double helix.
Until now you learned, that DNA encodes for amino acid sequences and I showed you how the DNA is organized. Now there is another problem which has to be solved: Who reads out the information stored on the DNA and how?
Well, the process of making proteins by using the information stored on the DNA is a two step one: in the first step (transcription) the DNA is read by an enzyme (a protein with a catalytic function, able to synthesize or degrade a certain molecule) called RNA polymerase and transcribed into a transport form called messenger RNA (RNA is a chemical very similar to DNA – the information coding properties are the same). This mRNA is then read by protein particles called ribosomes, which translate the DNA information into a protein (coding bases – amino acids). Therefore the second step is called translation. In many definitions, the part of genetic information that is necessary to produce one protein is called a gene. Besides the protein coding nucleotide sequence genes contain long nucleotide sequences which are in no way translated into proteins. These sequences are often regulatory sequences, responsible for the frequency a gene is transcribed and for the genetic regulation. A major fact is that the basic process of protein production is the same for all organisms in principle. This is why it is possible to take the genetic information of a plant or an animal and put it into a bacterium: The latter still can read the information, because all organisms use the same code and the same tools to read and recode the code into proteins.
Bacteria – general introduction
Everybody has heard of bacteria, for sure – maybe in connection with certain infectious diseases, with hygienic topics or with genetic engineering. And did you know, that there are ten times as many bacteria cells as human cells in the human body? But what exactly are bacteria? They are the most basal and most numerous organisms living on this planet. So you might wonder: Why have I never seen such a “bacteria”?. Well, the answer is simple: Bacteria are very small organisms. But what means small? Well, they are even a thousand times smaller than the smallest little point drawn on a piece of paper that you ever saw. If you placed one bacteria behind another you would need more than a million bacteria to get a line which is one feet long. They are only consisting of one cell, but can carry out all the major functions of life: they reproduce, they intake food and excrete metabolic wastes, they respire (convert food into energy) and they grow. And all this is done within a tiny cell of a few micrometers in length.
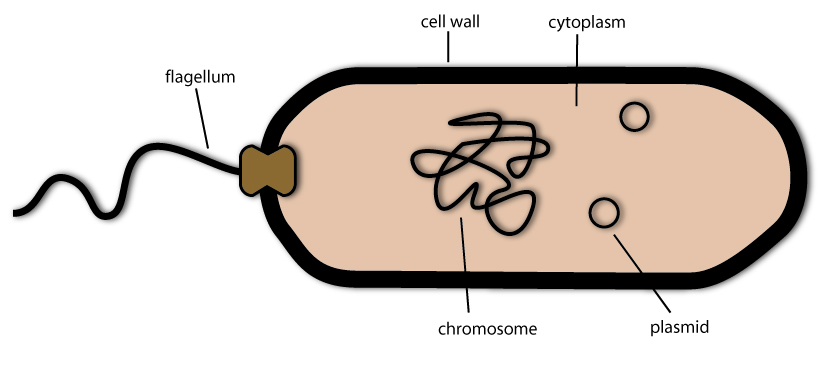
So how are these tiny living beings organized? Like every cell, they have an inner cytoplasm, which mostly consists of water and soluted proteins. This cytoplasm is the basis for all reactions carried out in the cell and provides an environment for molecule transport via diffusion. In the cytoplasm there lies the chromosome, which carries most of the genetic information of the bacterium. For example it encodes the proteins necessary for DNA replication, for the metabolism and for the secretion of substances. The bacterial chromosome lies free in the cytoplasm, unlike the ones in the cells of plants or animals - they are enclosed by an envelope and thereby forming the so called cell nucleus. Because bacteria cells do not have such a nucleus they are called procaryotes (the ones without a nucleus) - as opposed to the cells of plants and animals, which are called eucaryotes (the ones with a proper nucleus).
But unlike human cells, not all genetic information is included in the chromosome. Some lies in small circular units of DNA called plasmids. They function exactly like the chromosome, but they are smaller and some of them can be transported into other bacteria cells by a mechanism called conjugation.
Surrounding the cytoplasm is the plasma membrane – a thin layer of lipids, which separates the inside of the bacteria cell from the environment. On the outside of the plasma membrane there is the cell wall. Similar to the cell wall you may know from plant, this stiff construct gives the bacteria cell its shape and also shields the sensible plasma membrane.
Most bacteria are also able to swim. They do this via propeller like structures that are called flagella. These are long and thin protein fibers that rotate driven by a molecular motor. And they are not only able to tumble around randomly, but they can swim target-oriented in a process called chemotaxis.
The bacteria most well known to scientists are Escherichia coli (short version: E.coli) bacteria. They are organisms living in our lower intestine naturally - as a part of the normal flora of bacteria that live there. So most strains are harmless to humans, but there are some that can cause food poisoning. For most biotechnologists, E.coli bacteria are like their laboratory pets: They do most of their work on them and with their help. Because they are easy to grow and their genetics are relatively simple and easy to maipulate, E.coli is one of the best-studied model organisms in biotechnology.
Bacterial conjugation
Just imagine you would kiss somebody and thereby exchange genetic information with you mate, which helps you to survive under certain environmental conditions. Incredible? Well, this is exactly what bacteria cells are able to do and what is called conjugation!
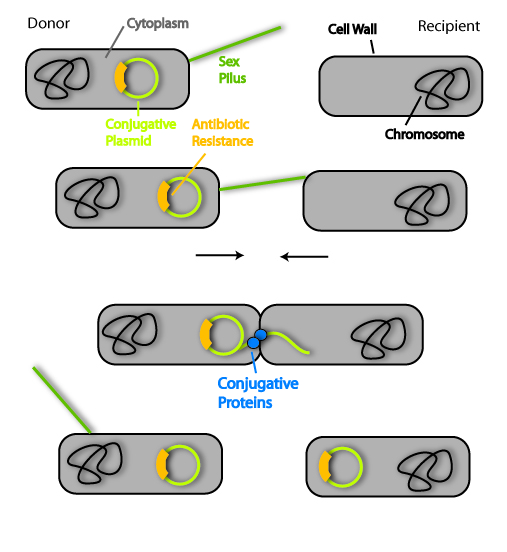
Scientifically, this term describes the transfer of genetic information from a donor bacterium to a recipient bacterium. They have to establish a cytoplasm bridge for this task, so the cells have to be in direct contact.
But how do the cells find each other and make this stable link? The donor cell has sensors to find and bind the recipient cell. These are protein extensions on the surface of the bacteria cell and they scan the surrounding area of the donor for recipients. Fittingly they are called sex pili. If these pili have established the contact between the two cells, they retract and bring the cells close together. The cytoplasm bridge is formed with the help of other protein factors.
In the next step, the genetic information is transferred from the donor to the recipient. Therefore another set of specialized proteins is needed. The genetic information generally is contained on plasmids, which are circles of DNA. As I said in the beginning, this information can be helpful to survive under special environmental conditions. One important example is antibiotic resistance. Normally, bacteria are killed by antibiotics. This is why they are used to fight infectious diseases evoked by pathogenic bacteria. But some bacteria have special proteins that allow them to survive even in an environment with antibiotics. And the information for these proteins can be transferred by conjugation.
You see, bacteria have an enormous plasticity as far as their genome is concerned and can therefore adapt to various conditions – due to the mechanism of conjugation.
Viruses
What is a virus? It is a sub microscopic particle – so we will never be able to see it with bare eyes. But we are able to feel the consequences of a virus acting – have you ever had a cold, the flu, herpes or chickenpox? These are diseases caused by viruses. They arise due to the lifestyle of a virus: These particles are not able to live on their own; they need a host cell to multiply. So you could call them cellular parasites. Therefore many scientists tend to not call them organisms, because they are not able to “live” in their own. By using the host cell for their own purpose, they often damage it severely – this is what causes the well known diseases.
There are not only viruses that infect humans, but there is a special virus for almost any kind of organism, for example there are a class of viruses who infect bacteria cells. They do the same to these cells as they do to human cells: They infect them, use their resources to multiply and in most cases destroy them to be set free. This is why they are called bacteriophages – bacteria eaters.
But how exactly does a virus/bacteriophages infection work? Therefore we have to know how a phage is organized: The core of the virus is its genetic information. This is wrapped in a coat of proteins, that shield it and also makes the contact to the host cell and injects the genetic information into this cell. There the information is copied and read by proteins of the host. In principle, the viral information tells the apparatus of the host cell, to make new viruses. So the virus programmes the host cell, e.g. the bacterium, to make more of this pathogenic agent that will kill it. Because the information of the virus also contains proteins, which degrade the cell wall of the bacteria and, in scientific terms, lyse it. That means the cell wall is not longer able to hold the pressure of the inner cytoplasm because it is tattered by the lysis proteins. The consequence: The bacteria cell bursts.Thereby the phages are set free and can look for other bacteria to infect.
Chemotaxis
The motility of bacteria is a quite interesting business. Even though bacteria are very small organisms they know quite well how to survive- and that means, in terms of biology, they want to chaw down every time at every date. But there is one problem with all good food in the environment: sometimes it is far away. And that means that bacteria need a functional motility system to be able to follow the direction where the smell of good, fresh food comes from. There is, of course, one important question: What food do bacteria like in nature? Bacteria eat everything you can imagine: They eat vegetarian food, meat, milk and many other things. Independent of the food being hot, cold or salty. Bacteria could be seen as the waste for all organic substances arousing in nature. But there is one meal they prefer: sugar. So bacteria are not so different from us... or don’t you like eating chocolate more than a tin of spinach?
Another aspect of bacterial survival is the detection of alarm substances showing the bacteria that they are close to something endangering their survival. Finding nutrient and avoiding conflicts and risks are basic prerequisites for the survival of organisms in nature. The system which is important for that is called Chemotaxis.
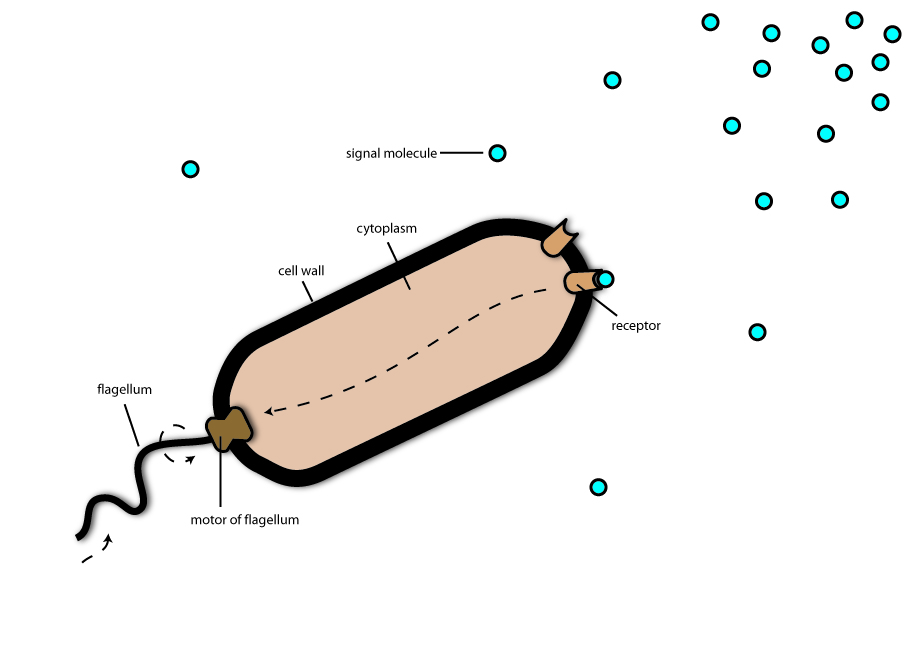
The chemotaxis system consists mainly of two different components: On component is the flagellum, which can be imagined as a rotating propeller. The interesting thing about this flagellum is, that it can rotate in two different directions: If the flagellum rotates in counter-clockwise direction the bacteria swims streight forward at high speed. In this state the flagellum rotates about 3.000 times per minute and accelerates the bacterium at a fulls speed of 50 µm (0.05 mm) per second- which is astonishing fast. You don’t believe that 50 µl/s is fast? Well, could you imagine to run at a speed of 170 mph? That is what bacteria do if you measure their speed in comparison to their length. But there is also a state, in which the bacteria flagellum rotates clockwise. In this state the bacteria is tumbling around and orientating in the environment. It could be compared to the sniffing of a dog, searching for a trail of the scent. The way the bacteria move, which is tumbling for a short time, then swimming forward a second and tumbling again is called the random walk.
In this state the second component of the chemotaxis system comes into game: the chemotaxis receptors. The chemotaxis receptors can be imagined as if they were little noses coming out of the bacterial body. Or like small grids for nutrients which means, that if the receptor catches a nutrient molecule it gets activated and sends an alert-signal to the bacteria: “Food”! Then the bacteria tumbles in the direction, where the receptor bound the nutrient melecule and begins to swim in this direction. As there are more and more nutrient molecules in the environment if the bacterium gets closer and closer to the food source it is able to find the right way just by following what we call the concentration gradient of the nutrient molecules. Just the opposite is the case, when bacteria detect alert substances. If you want to simulate chemotaxis at home just ask your friend or parents to hide something strongly and good smelling meal, i.e. pancakes or a piece of warm apple pie in your room and try to find it with shut eyes.
 "
"