Team:University of Lethbridge/Notebook/Project2October
From 2008.igem.org
Revision as of 23:40, 29 October 2008 by Munima.alam (Talk | contribs)
Back to The University of Lethbridge Main Notebook
Contents |
October 9, 2008
Roxanne, Munima, Christa, Sebastian
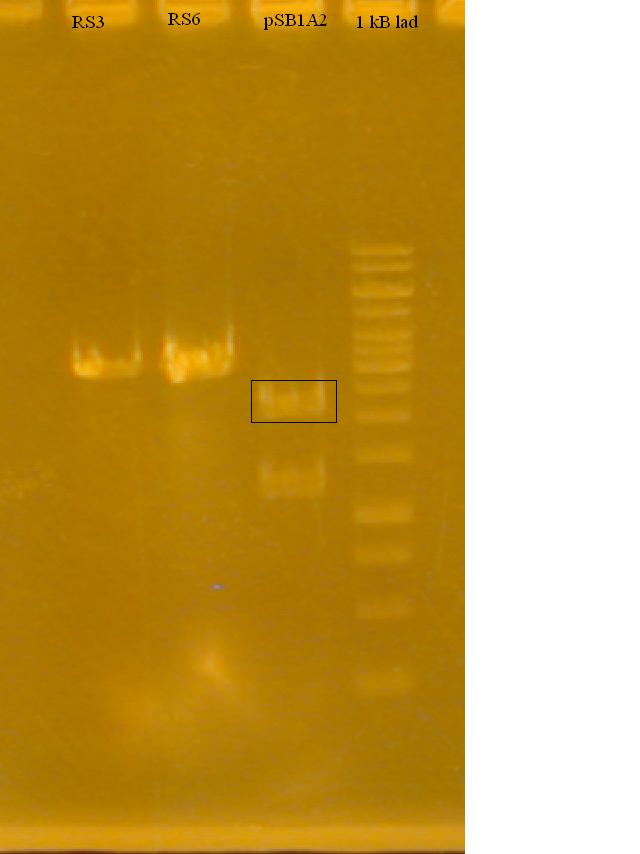
-Restriction digested RS3 and RS6 (produced by Nathan Puhl) and pSB1A2 + GFP sub with XbaI and SpeI
October 10, 2008
Roxanne
-Dephosphorylated pSB1A2 with Antarctic Phosphatase.
October 11, 2008
Roxanne
-Ran a gel to determine if the Restriction Enzymes cut the plasmid and RS3, RS6
October 12, 2008
Roxanne, Alix
-Restriction Digesting RS3 and RS6 again with XbaI and SpeI at 37.0 C for 2 hours.
-Attempting a new method of Gel Extraction (or rather an old method, new to us). Freeze 'n Squeeze.
Protocol: Freeze 'n Squeeze -Run the sample you wish to extract on a TAE-Agarose Gel -Cut out the band you wish to purify -Incubate in 1 gel volume 0.3 M NaCOOH at room temperature for 30 minutes. -Make your own spin column from a small microfuge tube with a hole cut out of the bottom, stuffed with glass wool. This tube should be inserted inside a 1.5 mL microfuge tube. -Transfer the solution to the spin column. -Freeze the tube in liquid Nitrogen for 1 minute, then spin at full speed for 15 minutes. -Precipitate the DNA in Ethanol. Remove the supernatant. -Wash in 75% Ethanol. Remove as much ethanol as possible. -Centrifige for 5 minutes then, remove the rest of the ethanol. -Let pellet to air dry for 10 minutes, this allows all ethanol to evaporate off. -Resuspend in TE Buffer, 10 uL. -Quantify either by Gel or UV Spec. -Proceed with ligation.
October 14, 2008
Roxanne
Objective: PCR of the riboswitch in a 50 uL x 9 reactions.
Master Mix (1x):
-10x Buffer: 5 uL -10 mM dNTP: 1 uL - 50 mM MgCl2: 1.5 uL - 10 mM reverse primer: 1 uL -10 mM forward primer: 1 uL -Taq polymerase: 0.2 uL -d2H2O: 39.3 uL
October 15, 2008
Roxanne
Another PCR of the riboswitch in a 20 uL x 9 reactions.
 "
"