Team:Calgary Wetware/Project
From 2008.igem.org
| Home | The Team | The Project | Notebook | Protocols | Results | Beyond the Lab | [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2008&group=Calgary_Wetware Parts] |
|---|
Contents |
Overall Project
Microorganisms use pheromones to monitor their own population density as well as to detect and interact with other microbial species in a process known as quorum sensing. For instance, bacteria can disrupt the pheromone signals of other competing bacteria, effectively preventing their proliferation. In a similar sense, we will exploit the natural communication systems involving Autoinducer-1 (AI-1) from Vibrio fischeri and Autoinducer-2 (AI-2) from Vibrio harveyi, to create a model biosensor system in Escherichia coli. We will engineer the genetic circuits necessary for the production of these pheromones into two populations of E. coli (termed Bad guy #1 and Bad guy #2, as per their respective Autoinducer). In addition, our third population of E. coli (termed Champion cell) acts as a biosensor by receiving these signal inputs and subsequently initiating transcription of specific E. coli-targeted bacteriocins (i.e. colicins). The presence of AI-1 induces the Champion to produce a colicin to which Bad guy #1 is susceptible, but to which Bad guy #2 is resistant, and vice-versa for AI-2. An additional aspect of our system is the ability of the Champion cell to report the presence of each specific Bad guy by producing a specific fluorescent protein (i.e. either green or red fluorescent protein) in tandem with the specific colicin as determined by the presence of either AI-1 or AI-2. We constructed this system using the molecular cloning methods utilized in iGEM. While the Vibrio fischeri components of our system were obtained as standardized parts from the iGEM Registry, we plan to clone and standardize all parts of Vibrio harveyi AI-2 quorum sensing system. All of the standardized parts will be flanked by the specific restriction endonuclease sites as required for iGEM BioBricks. This will allow for the simple and iterative directional cloning strategy for the construction of the necessary genetic circuits.
System #1 In Detail
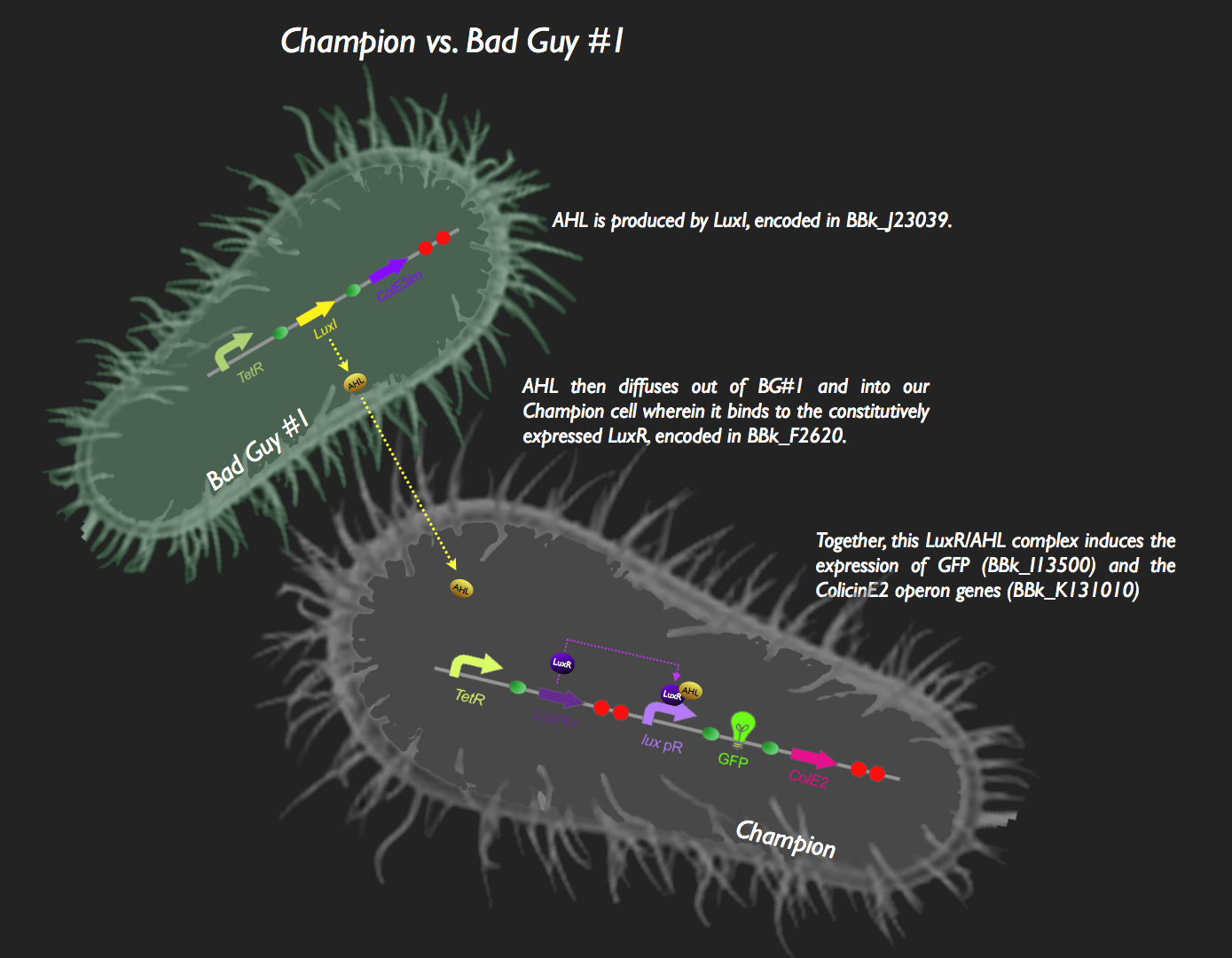
![]() The first half of our system has Bad Guy #1 as the culprit. Bad Guy #1 is capable of producing large amounts of autoinducer-1, more commonly known as Acyl Homoserine Lactone. This is through constitutive expression of the luxI gene. AHL begins to diffuse into our Champion Cell, which is constitutively expressing LuxR. LuxR protein binds to the pheromone, and the AHL-LuxR complex activates the lux pR promoter, leading to transcription of green fluorescent protein and the colicin E2 activity, immunity and lysis genes (CeaB, CeiB and CelB respectively). The champion cell now fluoresces green, and colicin expression results in the partial lysis of the cell but also release of CeaB, which would be taken up by Bad Guy #1. Colicin E2 would lead to the eventual destruction of Bad Guy #1, and our Champion Cell will rein supreme.
The first half of our system has Bad Guy #1 as the culprit. Bad Guy #1 is capable of producing large amounts of autoinducer-1, more commonly known as Acyl Homoserine Lactone. This is through constitutive expression of the luxI gene. AHL begins to diffuse into our Champion Cell, which is constitutively expressing LuxR. LuxR protein binds to the pheromone, and the AHL-LuxR complex activates the lux pR promoter, leading to transcription of green fluorescent protein and the colicin E2 activity, immunity and lysis genes (CeaB, CeiB and CelB respectively). The champion cell now fluoresces green, and colicin expression results in the partial lysis of the cell but also release of CeaB, which would be taken up by Bad Guy #1. Colicin E2 would lead to the eventual destruction of Bad Guy #1, and our Champion Cell will rein supreme.
This system was assembled in typical iGEM fashion. The part [http://partsregistry.org/Part:BBa_J23039 J23039] was transformed as is into E. coli to become Bad Guy #1. To construct the circuit in the Champion Cell, [http://partsregistry.org/Part:BBa_F2620 F2620], [http://partsregistry.org/Part:BBa_I13500 I13500] and the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K131009 colicin E2 operon] were constructed as shown in the picture. Colicin E2 was obtained from UC Berkeley, and PCR was used to biobrick it.
Tests were also performed, the results of which can be seen in the Results section. These were verifying that Bad Guy #1 is making AHL, verifying that our Champion Cell responds to AHL produced from Bad Guy #1, and that Colicin E2 was actually capable of preventing bacterial growth.
System #2 In Detail
![]() The other half of our system sees the aptly named Bad Guy #2 as the main adversary to our Champion Cell.
The other half of our system sees the aptly named Bad Guy #2 as the main adversary to our Champion Cell.
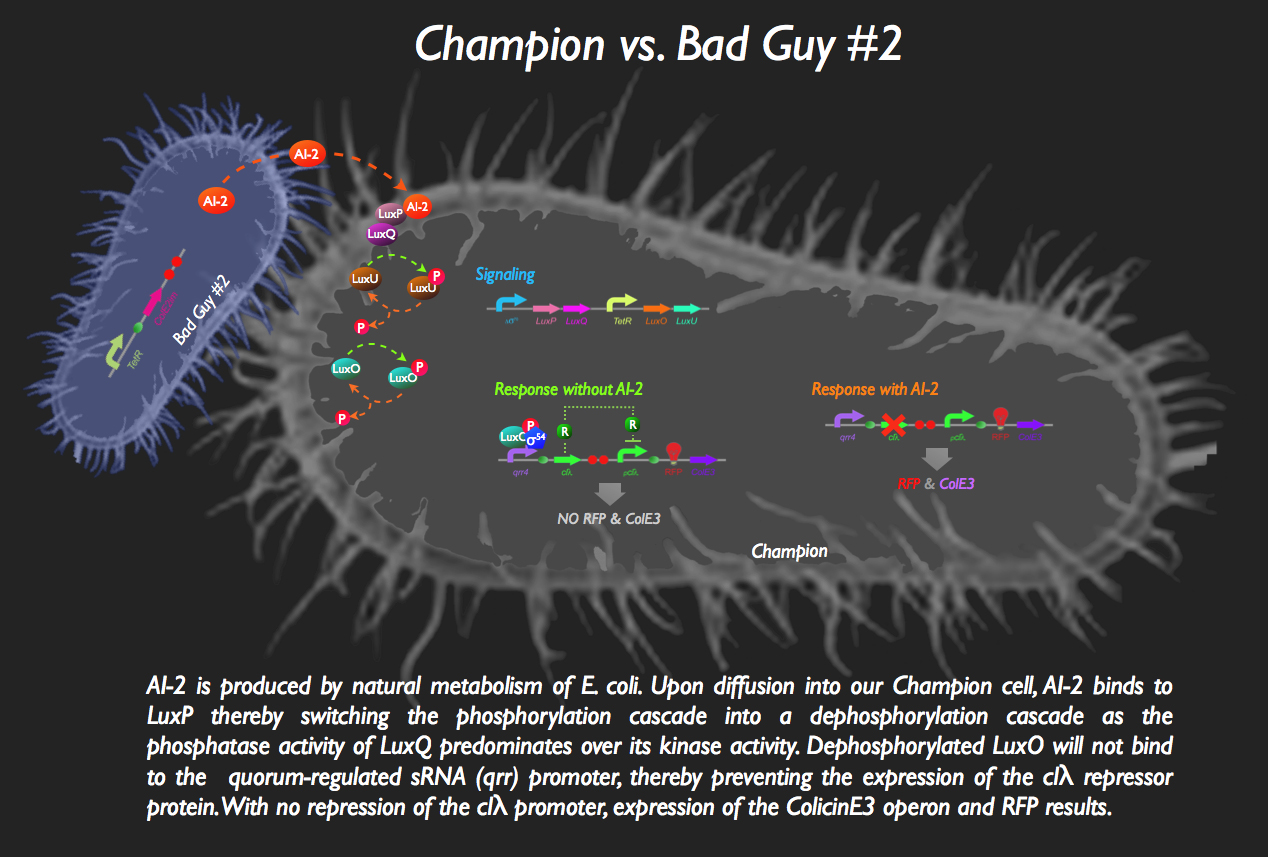
Without any AI-2 in the vicinity, however, there is downstream expression of a repressor protein, clλ, masking the expression of ColicinE3 and RFP:
But then bad Guy #2 comes along. He’s generating AI-2, which is bound by the periplasmic protein LuxP in the Champion Cell. The complex then binds to the inner membrane protein, LuxQ, which dephosphorylates LuxU. The loss of phosphorylation on LuxU means no more kinase activity, and LuxO spontaneously dephosphorylates. Since LuxO-p is required for the activation of the qrr4 promoter, no more clλ repressor is produced, activating the pclλ promoter. ColicinE3 is transcribed, leading to the eventual destruction of Bad Guy #2. Concurrently, our Champion Cell fluoresces red to inform the viewing gallery that the fight is going well.
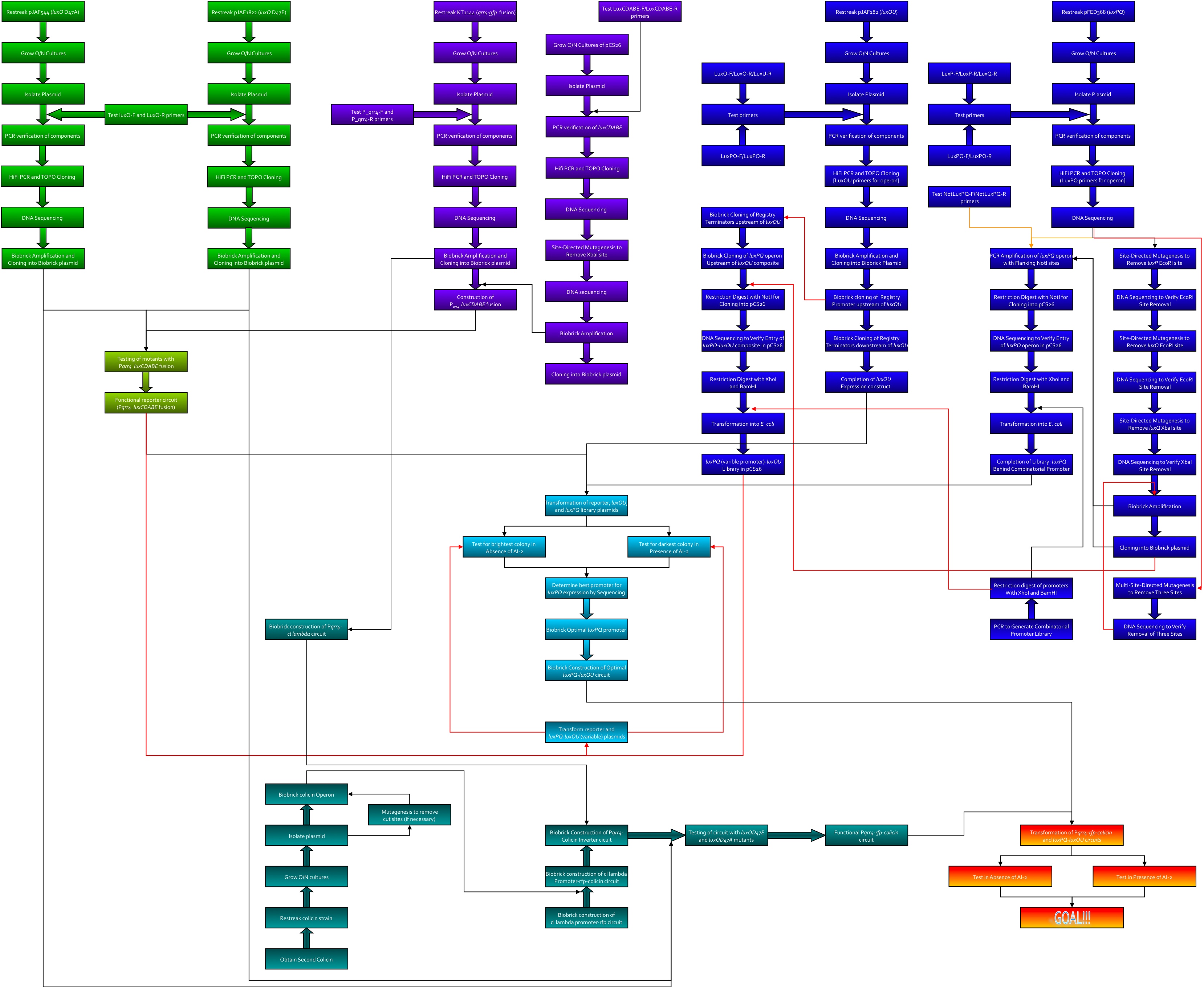
Here's the workflow envisioned for creating system 2. Kingsley worked on the green section, Kevin was to work on the purple, Thane worked on the blue, and Dave worked on the teal.
Applications
In essence, what’s proposed above is a model or proof of concept system. Then, where could this system be useful? One evident application stems from the knowledge that specific bacterial strains create bacteriocins targeted uniquely to their populations (i.e. E. Coli and colicins). Combining this bacteriocin with the AI-1 and AI-2 signaling pathways, the champion cell could effectively become a biosensor (i.e. fluorescing) and act as a living antibiotic towards actual ‘bad’ bacteria. For example, a bacteriocin against Salmonella strains could be engineered within our system in the place of the colicin genes. Then, when faced with an outbreak of Salmonella typhi, for instance, the champion cell could be deployed to detect contaminated foodstuffs, as well as eliminating the S. typhi. While this may sound unlikely, there was actually a paper published recently using a similar system of live E coli to help prevent AIDS transmission. And while that may sound just as far-fetched, the system is currently in the clinical trial phase.
 "
"