Team:MIT/Project
From 2008.igem.org
| Home | The Team | The Project | Experiments | Parts Submitted to the Registry | Results | Notebook |
|---|
Overall project
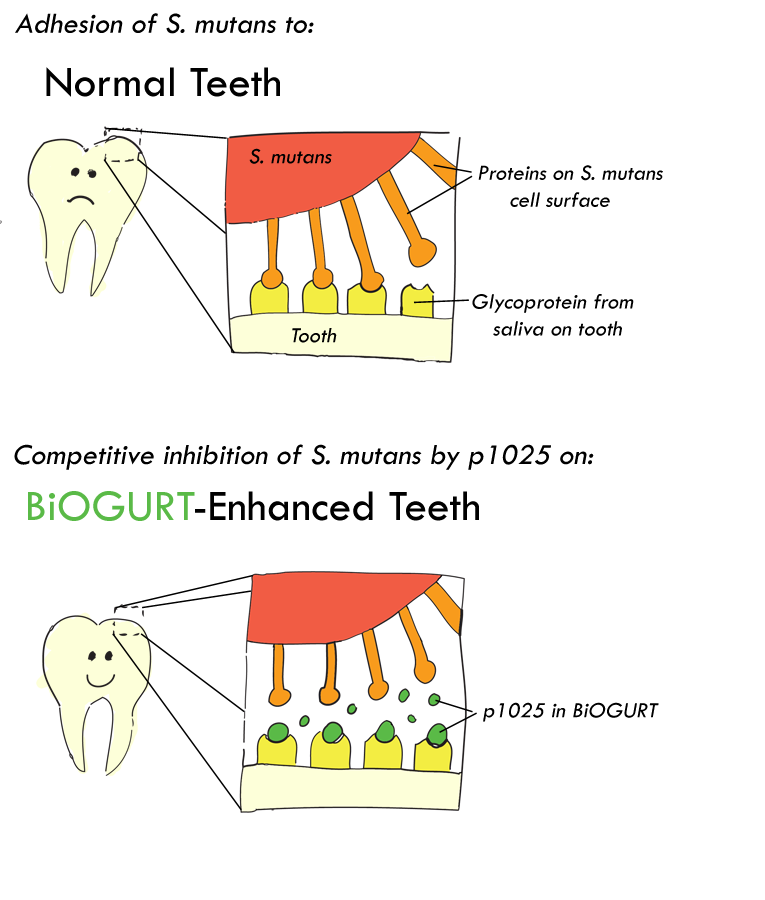
A clinical study (Kelly CG et al.; Nature Biotechnol. 1999) reports that a short 20 amino acid synthetic peptide called p1025 can reduce oral colonization of a major tooth-decaying bacterium Streptococcus mutans and thus help to maintain oral health. The peptide does so by competitively inhibit binding of S. mutans to a glycoprotein from saliva on tooth surface. The peptide is considered advantageous because it selectively prevents S. mutans colonization without removing beneficial bacteria that are also present in the mouth. In fact, the removal of S.mutans allows other (unharmful) bacteria to colonize these regions. It was found that introduction of p1025 into the mouth can inhibit S.mutans colonization for up to about 90 days.
Our research goal is to engineer one of the common yogurt bacteria, Lactobacillus bulgaricus, to secrets p1025 in yogurt. Consumption of this yogurt after a meal could replace teeth brushing and reduce tooth decay significantly (although it wouldn't help with bad breath!)
We are not going to market this yogurt. However, our project will demonstrate a bio-engineering approach to increase the value of common probiotics. Engineering yogurt is especially exciting. A new batch of yogurt is made by taking a small sample of the previous batch and using that to provide the bacteria for the new batch. This means that a starting culture of engineered L.bulgaricus can be used to provide a continuous supply of teeth cleaning yogurt. Since the p1025 gene could be replaced by any other gene or sequence of genes, any number of useful yogurt strains could be made. We believe that this could be the key to necessary, sustainable, and effective health care in small-scale underdeveloped countries, especially if yogurt is already an integral part of the diet.
Project Details
Stage 1
- Construction of p1025 fusion peptide with appropriate markers and expression of gene in E. coli. This is an intermediate step to evaluate gene construction and protein efficacy
Stage 2
- Binding Assay - To reproduce the results of the Nature paper and test if the p1025 construct made in stage 1 inhibits binding of Streptococcus mutans to hydroxyapatite beads as well as the original p1025 peptide.
Stage 3
- Expression of p1025 in Lactobacillus bulgaricus
Stage 4
- Make yogurt with modified Lactobacillus and test for inhibition of S. mutants binding
The Experiments
- For daily information about wet-lab experiments performed, please follow directions in the "Procedural Notebook section" of [[1]]
- For any further details regarding experiments, please refer to the appropriate sub-sections of [[2]]
A Visual Respresentation
| Home | The Team | The Project | Experiments | Parts Submitted to the Registry | Modeling | Notebook |
|---|
 "
"