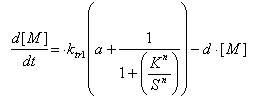Team:NTU-Singapore/Modelling/Constructs
From 2008.igem.org
|
Contents |
Constructs of the system
The system can be viewed as two parts. The first part comprises of lactose induced production of colicin E7 and the immunity protein. The second part comprises of a detection mechanism that produces the lysis protein upon the detection of both Iron ions and Ai-2 ( Autoinducer 2).
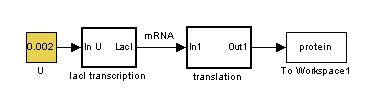
Constitutive Protein Synthesis
| 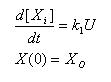
| 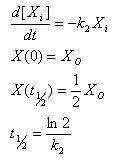
|
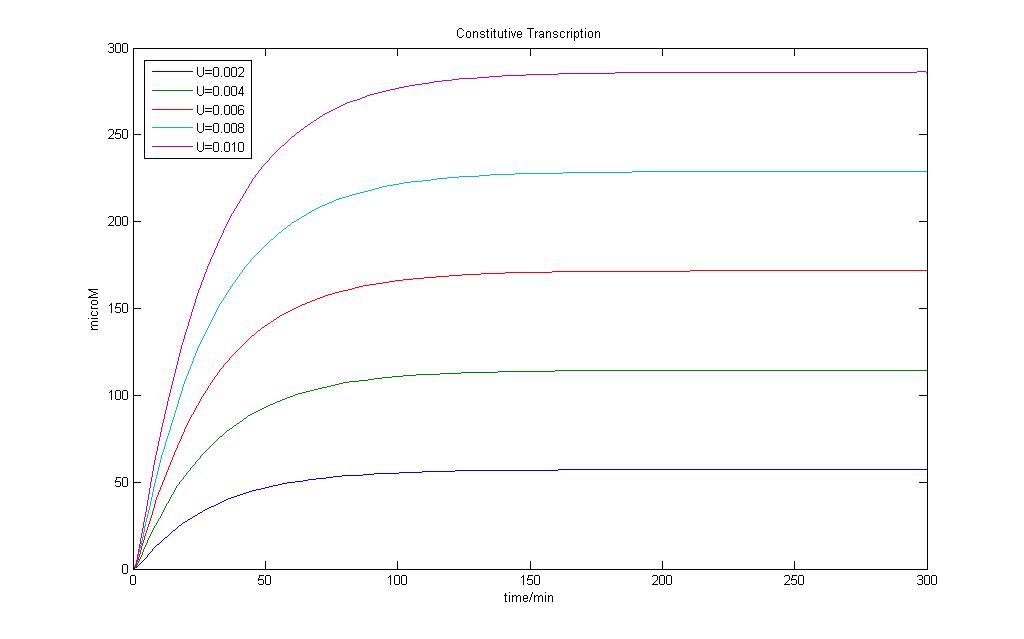
The constitutive model allows us to understand how fast steady state can be achieved for basal transcription.
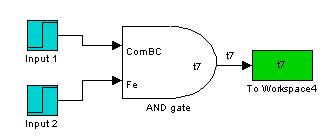
AND Gate
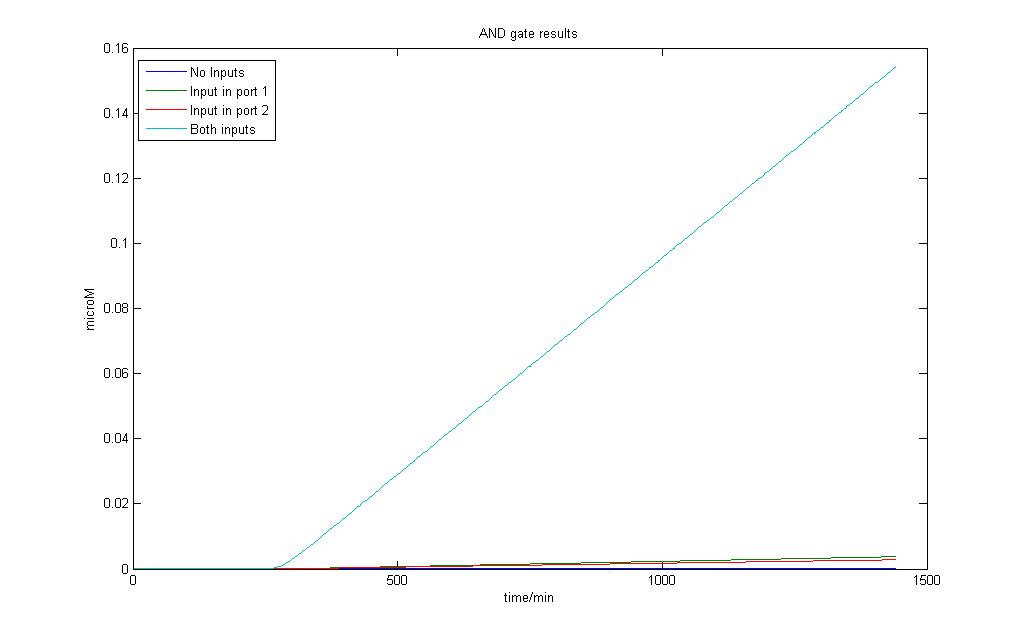
Addition of inputs was done at 250 minutes.
The AND gate model allows us to observe that even when there is no input, basal transcription still yields a certain amount of product. The difference is observed both inputs are applied and a sharp increase over the basal amount can be easily observed.
We can observe AND gate behaviour in this sense as it is impossible to have no protein production at all even in the most well-regulated system.
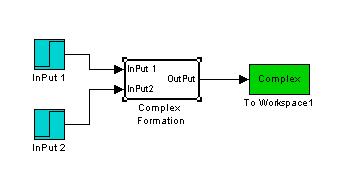
Complex Formation
This is the Simulink Model Construct for Complex Formations. The model is tested by applying an input when time = 150 min.
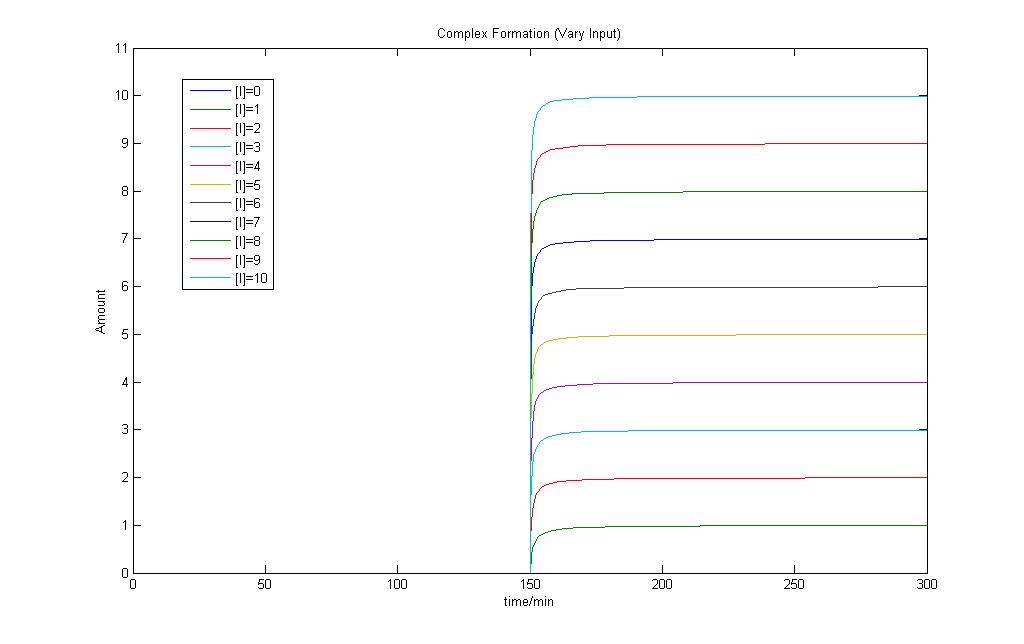
By increasing the input, we can see an increase in the amount of complex formed.
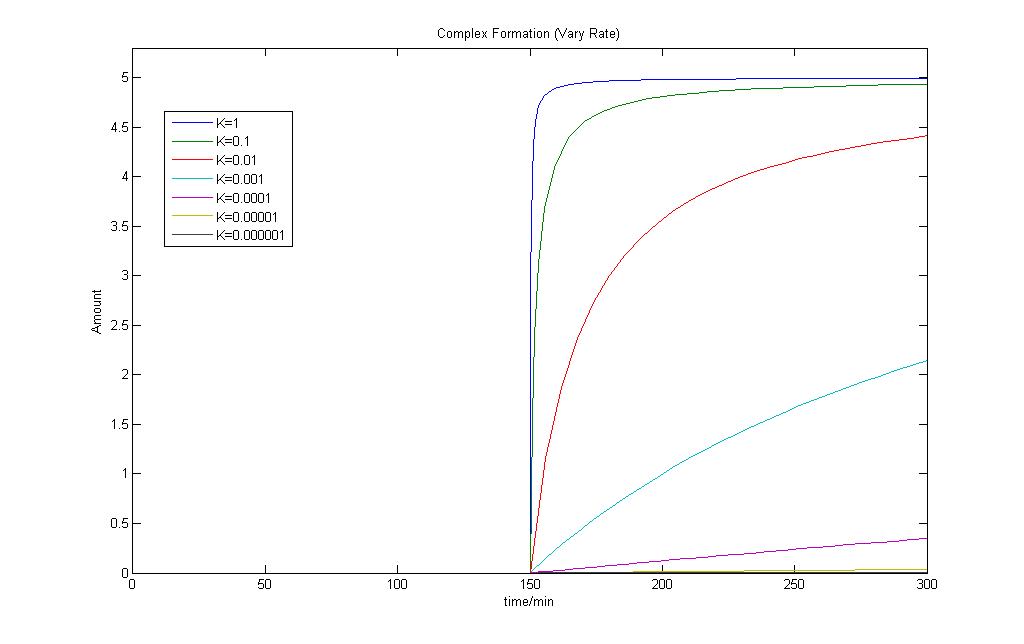
By changing the rate of Complex formation, we can also see a change in the amount of complex formed.
Both results show that the model works fine and can be used in our modeling exercises later on.
Regulated Transcription
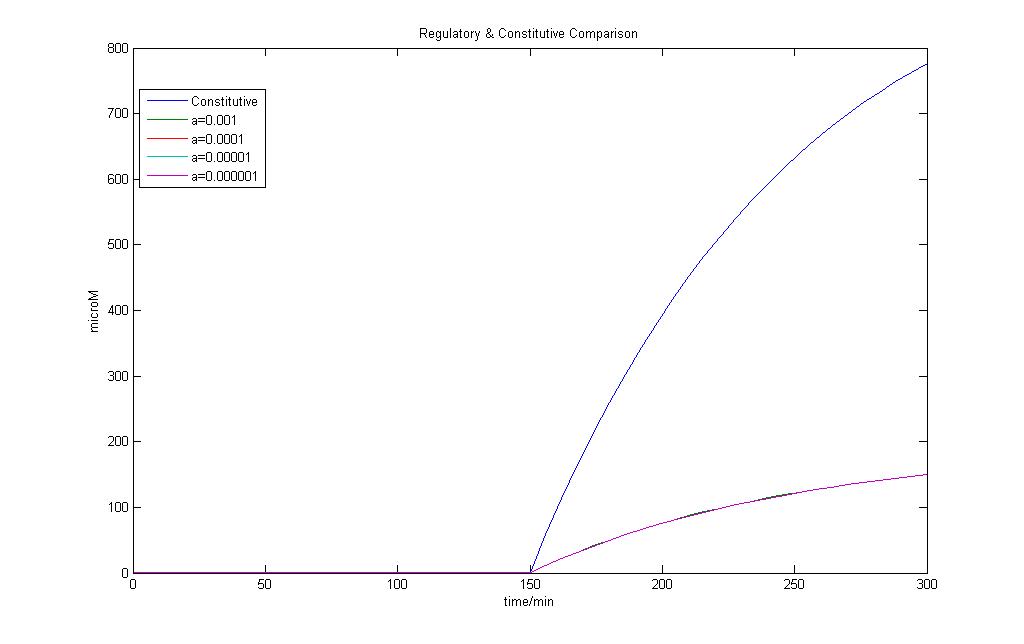
The model for regulated transcription is compared with the model used in Constitutive transcription. Input into both models was done when time = 150 min. This would allow us to see if there is a difference under two different conditions for the two models.
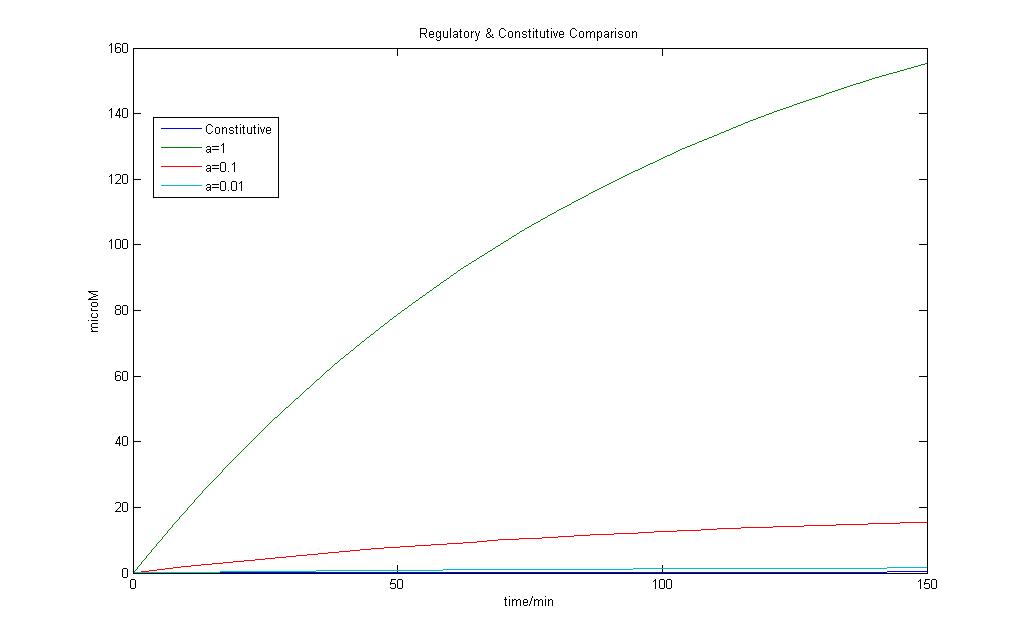
The results of the modeling exercise shows that if the parameter a > 0.01, it would introduce a basal rate transcription that is higher than that of constitutive transcription.
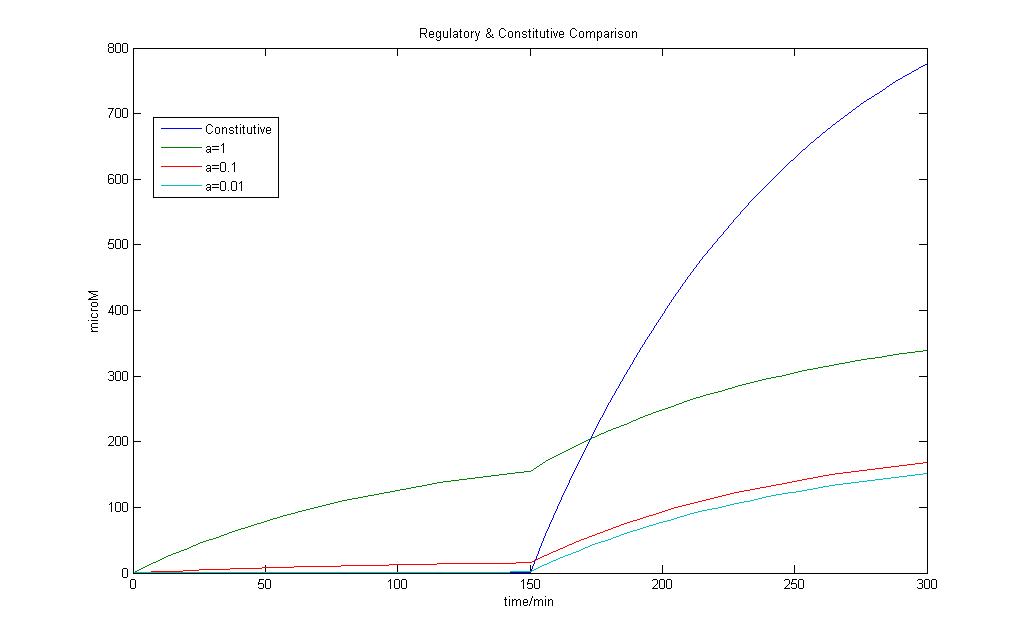
However upon the introduction of a stronger signal, constitutive transcription is still faster than that of regulated transcription.
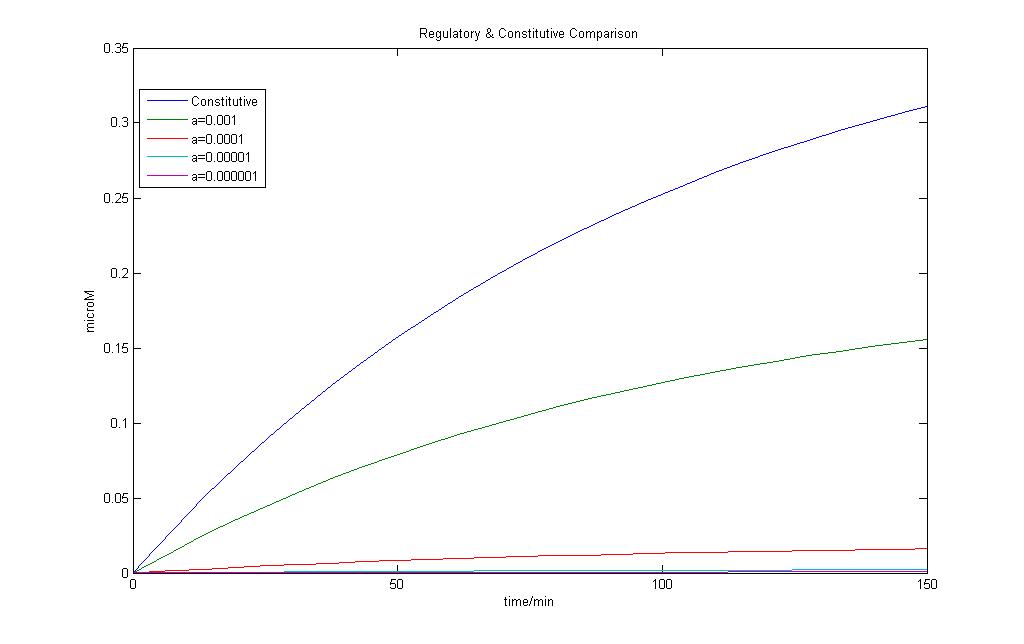
If parameter a < 0.01, we can see that constitutive transcription is faster than that of regulated transcription. The formation of mRNA is slower and more strongly regulated.
 "
"