Team:Paris/Modeling/BOB
From 2008.igem.org
Introduction
BibliographyMuch of our inspiration comes from four articles to which we shall refer in the next subsections :
Steps modeledHere is a quick summary of the step we decided to interpret mathematically:
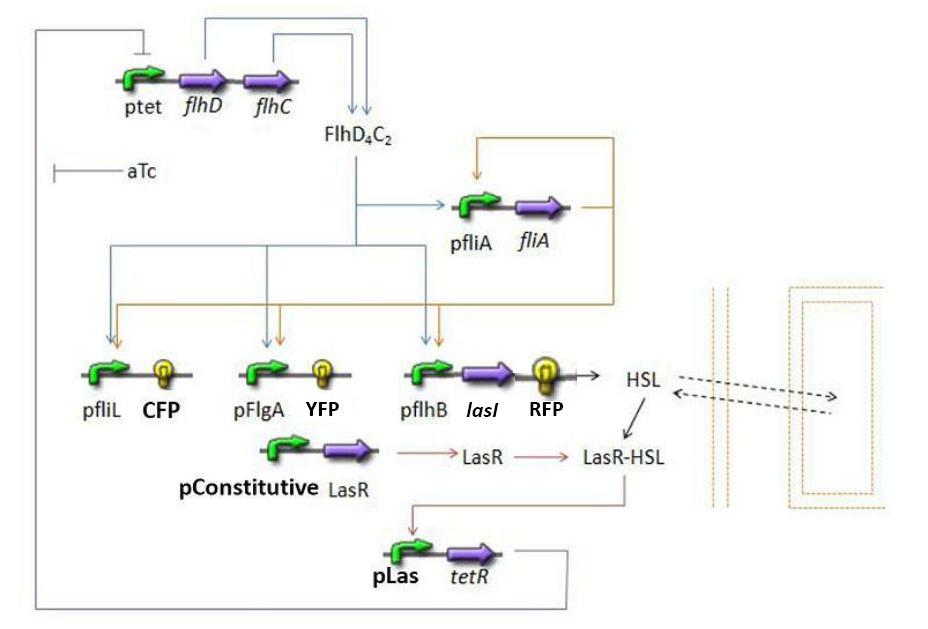
Using the Las system:
Or the Lux system:
Establishing the modelPopulation evolutionFirst of all, it seems important at this stage to present the way we interpret the evolution of the population in the chemostat. We want to impose to our model the fact that the rate of production has to be proportional to the existing population and to the amount of available resources. Hence a logistic model of the population, where c denotes the concentration of cells in the medium: Then, we need to add the death term, and a dilution term cause by the renewal of the chemostat. Finally we get : Where cmax is the carrying capacity for cell growth, and D the dilution rate, d the death rate constant FIFO Temporal Order ModuleInformations about the FIFO Temporal Order module Steps involved
Mathematical modelClassically we use the following equation to model gene interactions : where [Y] denotes the concentration of Y protein and γ its degradation rate (which unit is time-1).
β and β’ represent the relative influence of flhDC and fliA respectively, the units of β and β’ being time-1.
DiscussionNormalizationWhich gene goes were?Simulations and ResultsSynchronization ModuleUsing Las systemSteps involvedMathematical ModelDynamics of HSLDynamics of HSLextTetR=f(HSL)FlhDC=f(TetR)Using Lux systemfliA = f(FlhDC,FliA) (1) RFP and lasI behind pFliL = f(FlhDC,FliA) (2) Influence of lasI over HSL (3) Relation between HSL and HSLext (4) Influence of HSL over tetR (5) flhDC = f(tetR) (6) 4.3.2 Mathematical model First of all, we naturally keep the same model for steps (1) and (2). 4.3.2.1 Dynamics of HSL First and foremost, HSL is the key element in the synchronization process. We used a diffusive specie in order to balance the concentrations between cells. The outside medium concentration becomes an intermediate variable. We ended with
Where is the diffusion rate expressed in time-1 and
Then, we had two choices to model this step. At first we were inspired by [2] where the production of HSLint is described as being linear, and we were given the value of the constant. However, the results presented in this study were introduced as only being theoretical. The degradation rate comes in quite naturally.
We chose to model the creation of HSL by a Michaelis Menten expression. In fact, we found out that lasI seemed to be a monomeric enzyme, thus setting the Hill parameter to 1.
Here, is expressed in the concentration unit, and in the concentration unit by time unit. It is important to note that we took into account the fact that lasR/(lasR linked to HSL) was a function of HSL. However, we assumed that the fraction of HSL bound to lasR would not influence HSL concentration; this is the reason why, no “degradation due to binding term” appears in the HSL model. 4.3.2.2 Dynamics of HSLext The evolution of HSLext may be described with three terms. First of all, there is a dilution rate, Drenewal, due to the renewal of the chemostat. There is the usual degradation rate, .
Which gives
4.3.2.3 TetR = f(HSL) Here we had the choice to model the complexation phase of HSL and lasR, and then the binding phase of this complex on the pLas promoter. We assumed that those stages were short, and then decided to model them by a single step.
We checked in the registry that the pLas sequence was the same as in [promoter specificity]. It is explained that there are two operators, denoted OP1 and OP2. The Hill parameters were detailed in this article, hence our modeling choice :
4.3.2.4 FlhDC = f(TetR) For this last step, since we have not found any article indicating that we could simplify the model by using a linear function. Then, we chose a complete Hill function to describe the dynamics of flhDC:
4.3.3 Experiment and discussion Parameters summaryResultsOscillationsFIFOSynchronization |
 "
"