Team:KULeuven/Model/Output
From 2008.igem.org
Contents |
Output
Position in the system
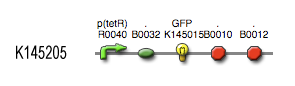
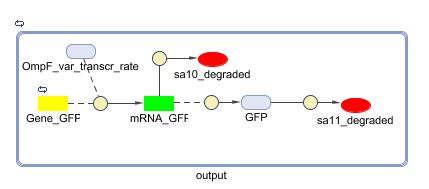
The output system is one of three modules directly linked to the input. The output system is a simple gene regulation, of which transcription is activated by tetR. The output signal is GFP (green fluorescent protein).
In the applied project of dealing with Crohn's disease this would be replaced by the drug necessary to cure the local inflammation. The drug concentration would be proportional to the amount of local inflammation sensed.
Describing the system
see also: Project:Output
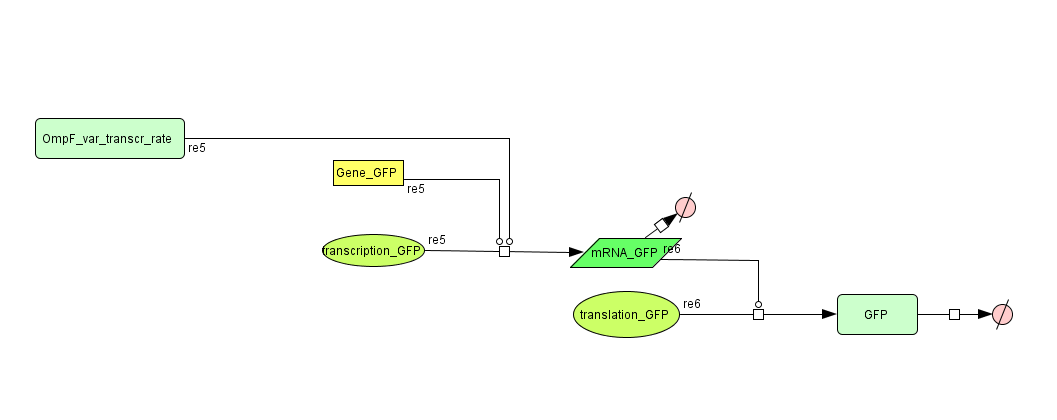
ODE's
Parameters
| Name | Value | Comments | Reference |
|---|---|---|---|
| Degradation Rates | |||
| dGFP | dLVA = 2.814E-4 s-1 | LVA-tag reduces lifetime to 40 minutes | (1) (4) |
| dmRNA_GFP | 0.0023 s-1 | (2) | |
| Transcription Rates | |||
| TetR_var_transcr_rate | p(TetR) dependent | (GFP) between 5E-5 and 0.0125 s-1 ~ [aTc] | (3) |
| Translation Rates | |||
| kGFP | 0.167 s-1 | translation rate for B0032 RBS (0.3 relative efficiency) | (5) |
Models
CellDesigner (SBML file)
Matlab (SBML file)
Simulations
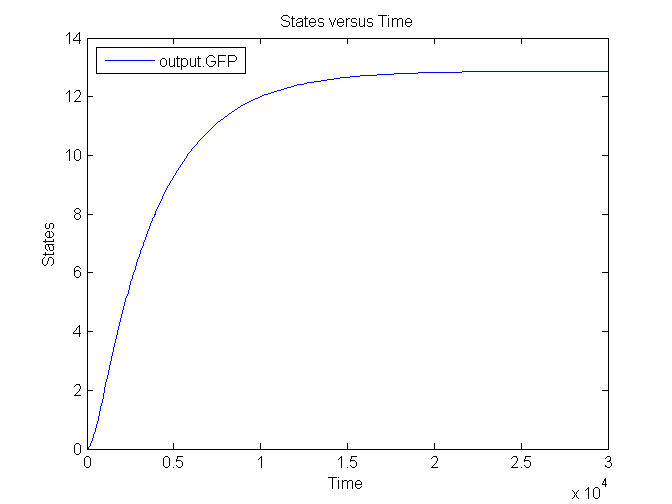
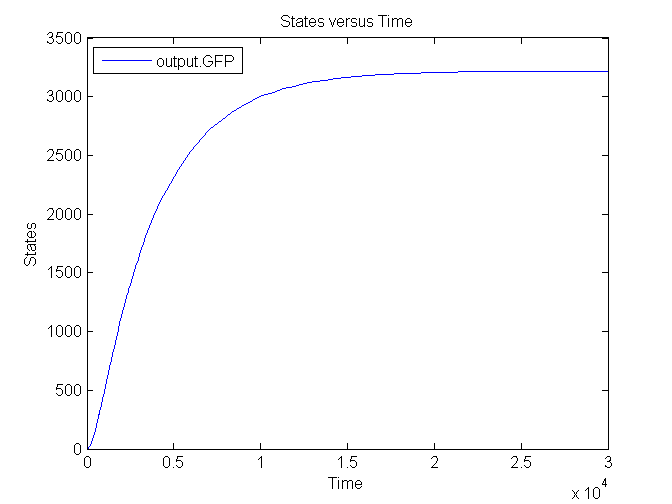
Several simulations were conducted, the following two show the transient respons and eventual saturation of the output system to a low and a high step signal as input, respectively. We see that there is a great distinction in the number of CFP molecules between the two input signals.
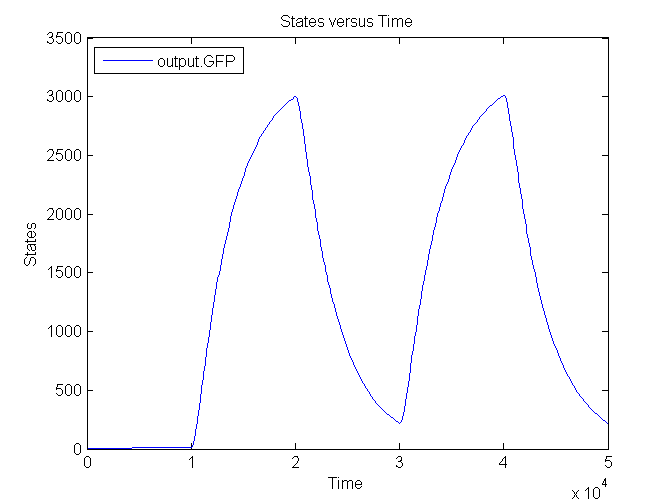
Next figure shows the switching behaviour of the output system to an light signal turning off and on:
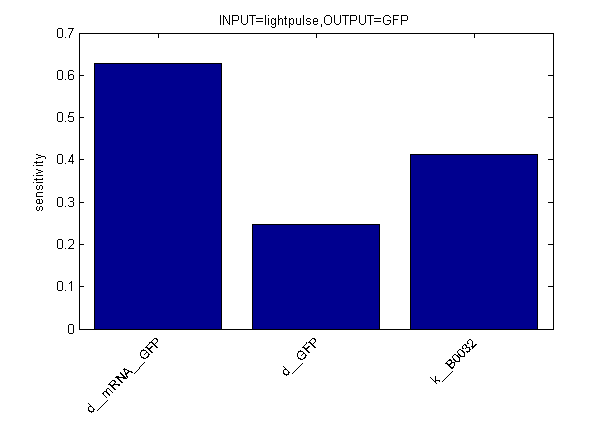
Sensitivity Analysis
References
| [1] | “Berkeley2006-RiboregulatorsMain - IGEM”; http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain. |
| [2] | F.J. Isaacs et al., “Engineered riboregulators enable post-transcriptional control of gene expression,” Nat Biotech, vol. 22, Jul. 2004, pp. 841-847. |
| [3] | J.A. Bernstein et al., “Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays,” Proceedings of the National Academy of Sciences of the United States of America, vol. 99, Jul. 2002, pp. 9697–9702. |
| [4] | “IGEM:Tsinghua/2007/Projects/RAP - OpenWetWare”; http://www.openwetware.org/wiki/IGEM:Tsinghua/2007/Projects/RAP#Model_and_simulation. |
| [5] | M. Gonzalez et al., “Lon-mediated proteolysis of the Escherichia coli UmuD mutagenesis protein: in vitro degradation and identification of residues required for proteolysis,” Genes Dev., vol. 12, Dec. 1998, pp. 3889-3899. |
| [6] | M. Bon, S.J. McGowan, and P.R. Cook, “Many expressed genes in bacteria and yeast are transcribed only once per cell cycle,” FASEB J., vol. 20, Aug. 2006, pp. 1721-1723. |
| [7] | “Part:BBa J23109 - partsregistry.org”; http://partsregistry.org/Part:BBa_J23109. |
| [8] | J.P. McDonald et al., “Regulation of UmuD cleavage: role of the amino-terminal tail,” Journal of Molecular Biology, vol. 282, Oct. 1998, pp. 721-730. |
 "
"