Team:UNIPV-Pavia/Project
From 2008.igem.org
Contents |
Overall project
We are trying to mimic Multiplexer (Mux) and Demultiplexer (Demux) logic functions in E. coli.
In the following paragraphs project details will be described from both digital electronic and genetic points of view.
Electronic Implementation
What kind of components are Mux and Demux?
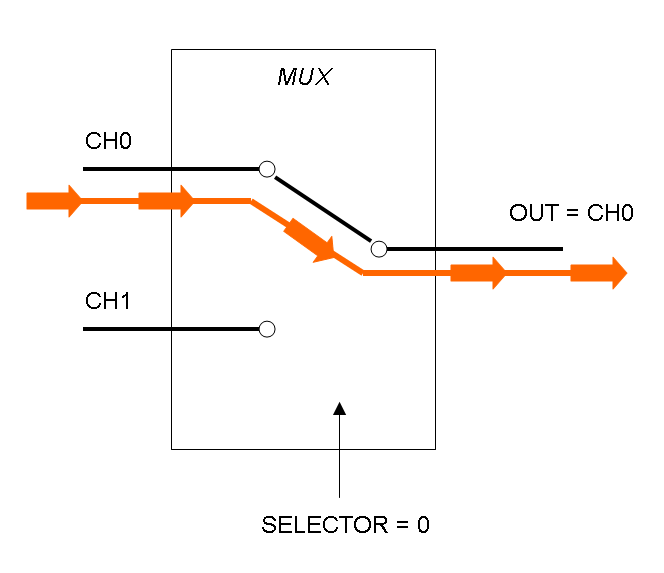
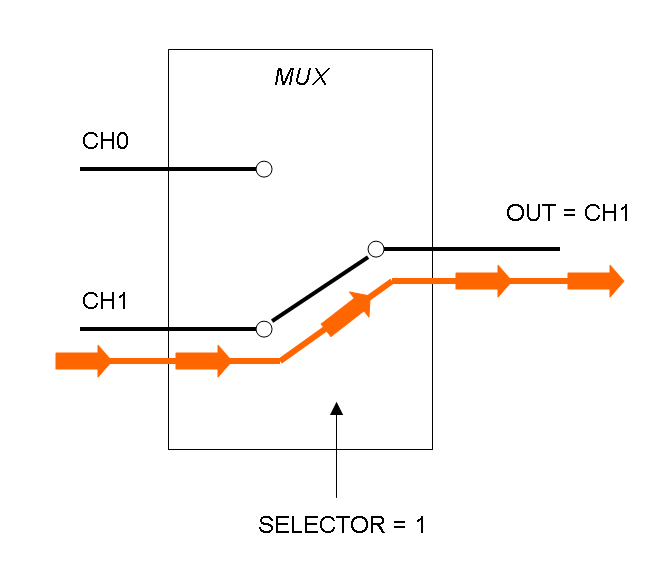
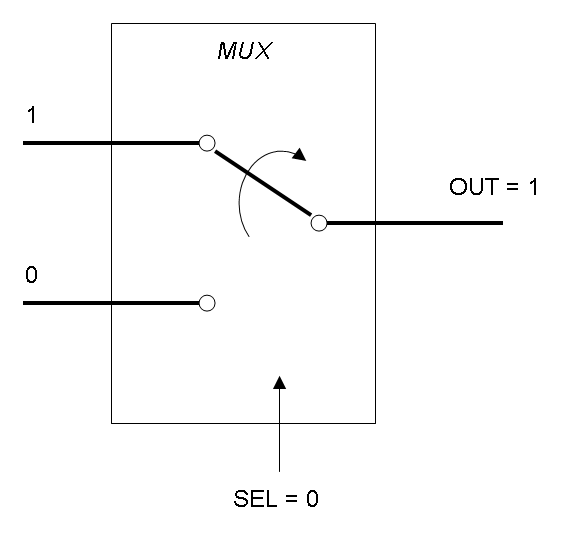
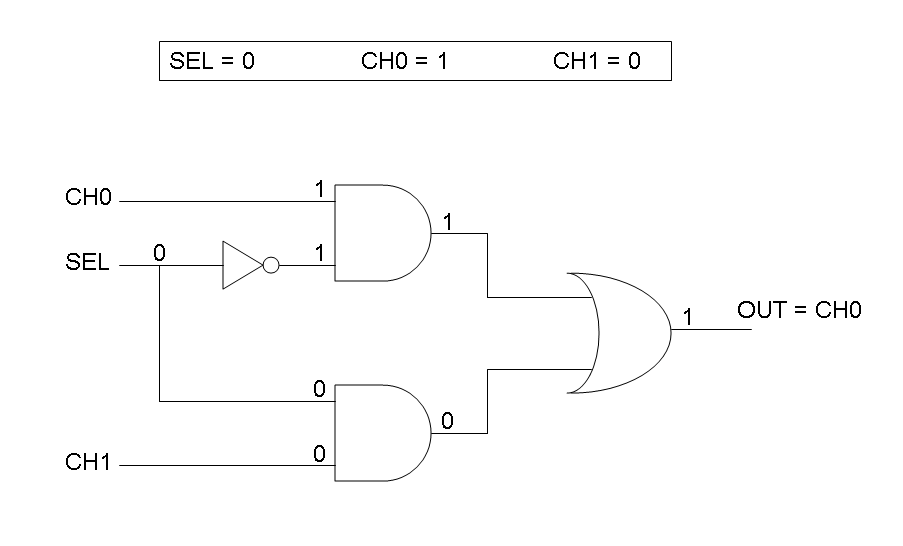
Mux is a component which conveys one of the two input channels values into a single output channel. The choice of the input channel is made by a selector.
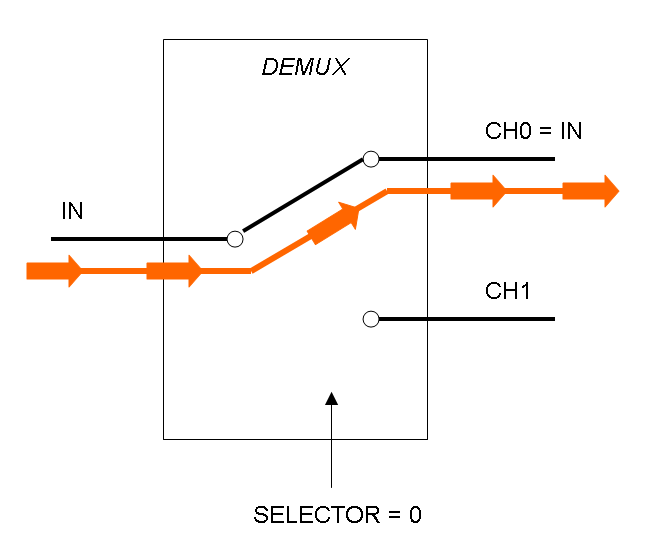
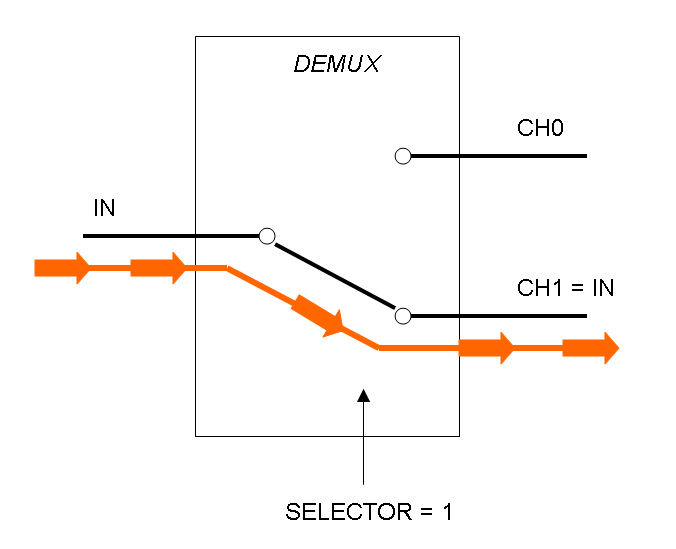
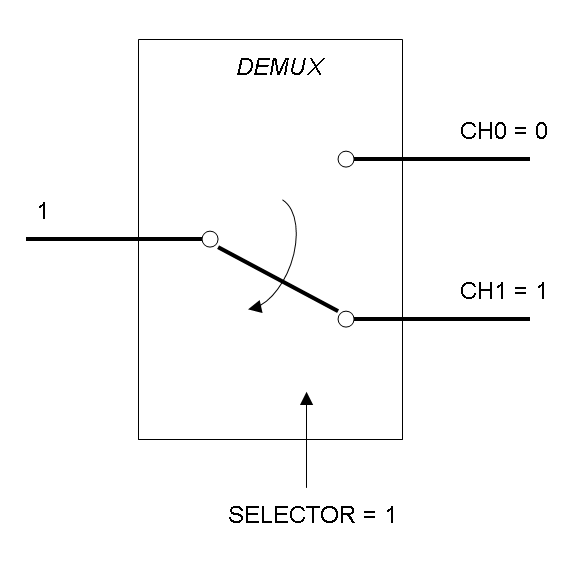
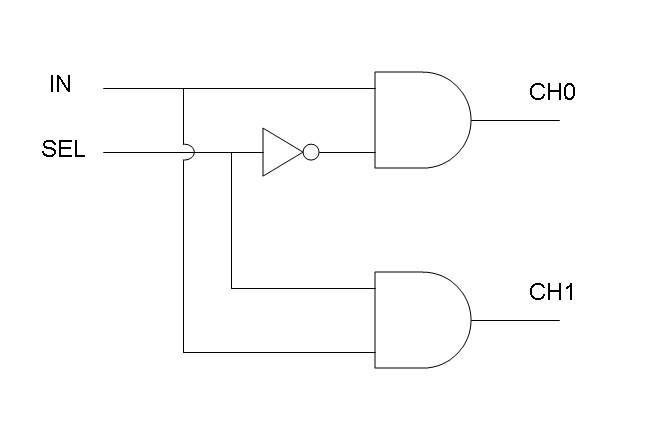
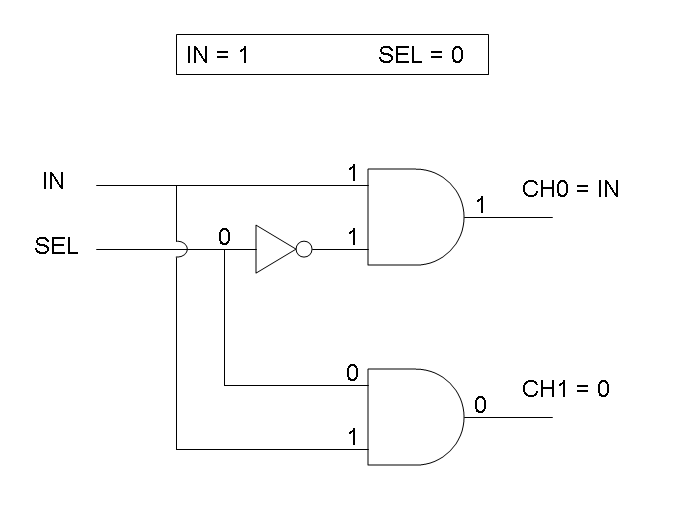
Demux is a component which conveys the only input channel value into one of the two output channels. The choice of the output channel is made by a selector.
The following pictures show data flow in Mux and Demux:
What kind of signals do we process?
In this project we consider Boolean logic signals, thus every input/output value can assume only the values 0 and 1. A function that processes Boolean values is called logic function.
Mux and Demux can be considered by now as black boxes which implement a logic function that can process input signals to output signals. Here you can see examples of Boolean data flow in Mux and Demux:
In the following documentation we will see what is inside these black boxes.
How can we formalize Mux and Demux logic behavior?
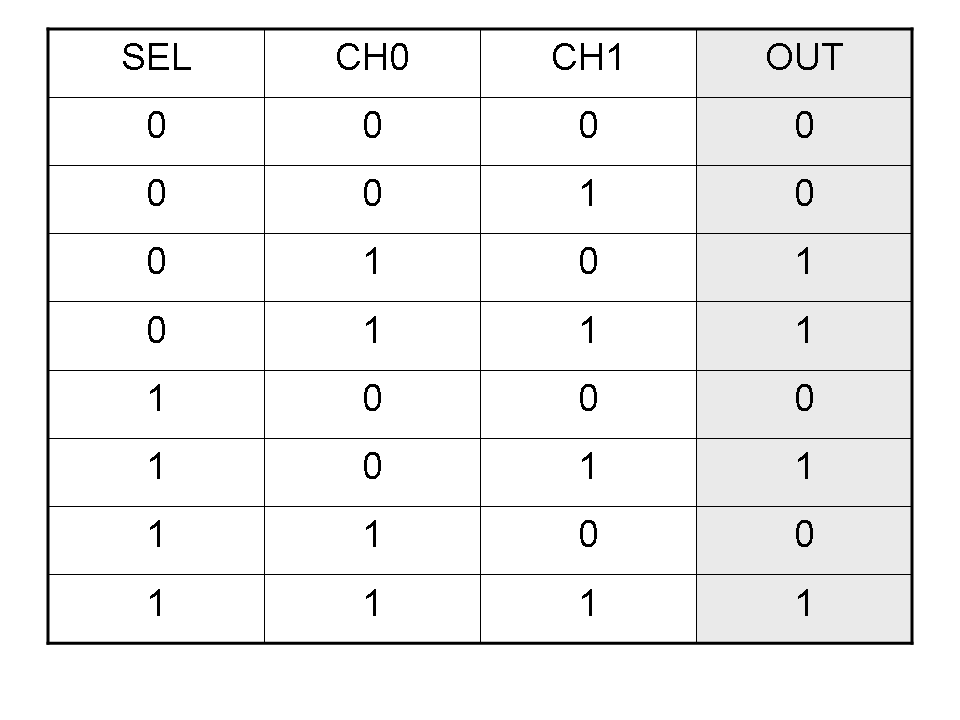
Logic functions can be formalized writing a truth table; a truth table is a mathematical table in which every row represents a combination of input values and its respective output values. The table has to be filled with every input combination.
Here you can see Mux and Demux truth tables (output columns are gray):
Building a logic circuit from a truth table
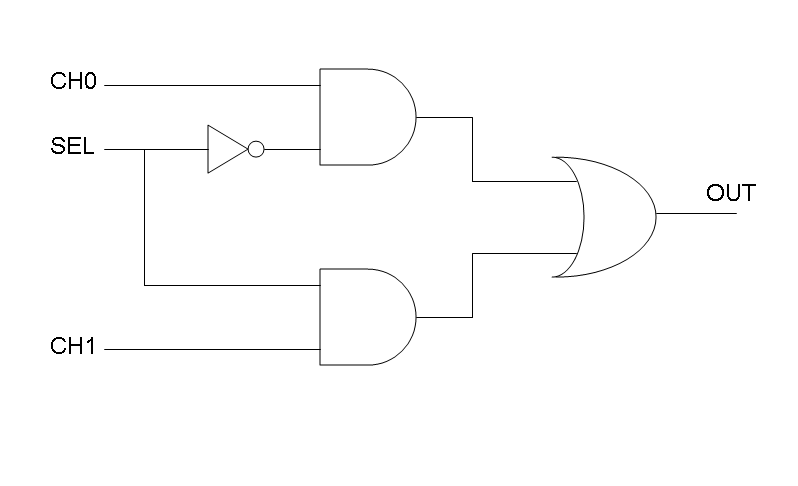
Our goal in this section is to project two logic gates networks which behave like Mux and Demux truth tables. A very useful tool to transform a truth table into a logic network is Karnaugh map.
It is possible to read about Karnaugh maps at: [http://en.wikipedia.org/wiki/Karnaugh_map]
Following Karnaugh maps method, we can write these two logic networks for Mux and Demux:
Genetic Implementation
Our goal is to mimic Mux and Demux logic networks in a biological device, such as E. coli. To perform this, we use protein/DNA and protein/protein interactions to build up biological logic gates.
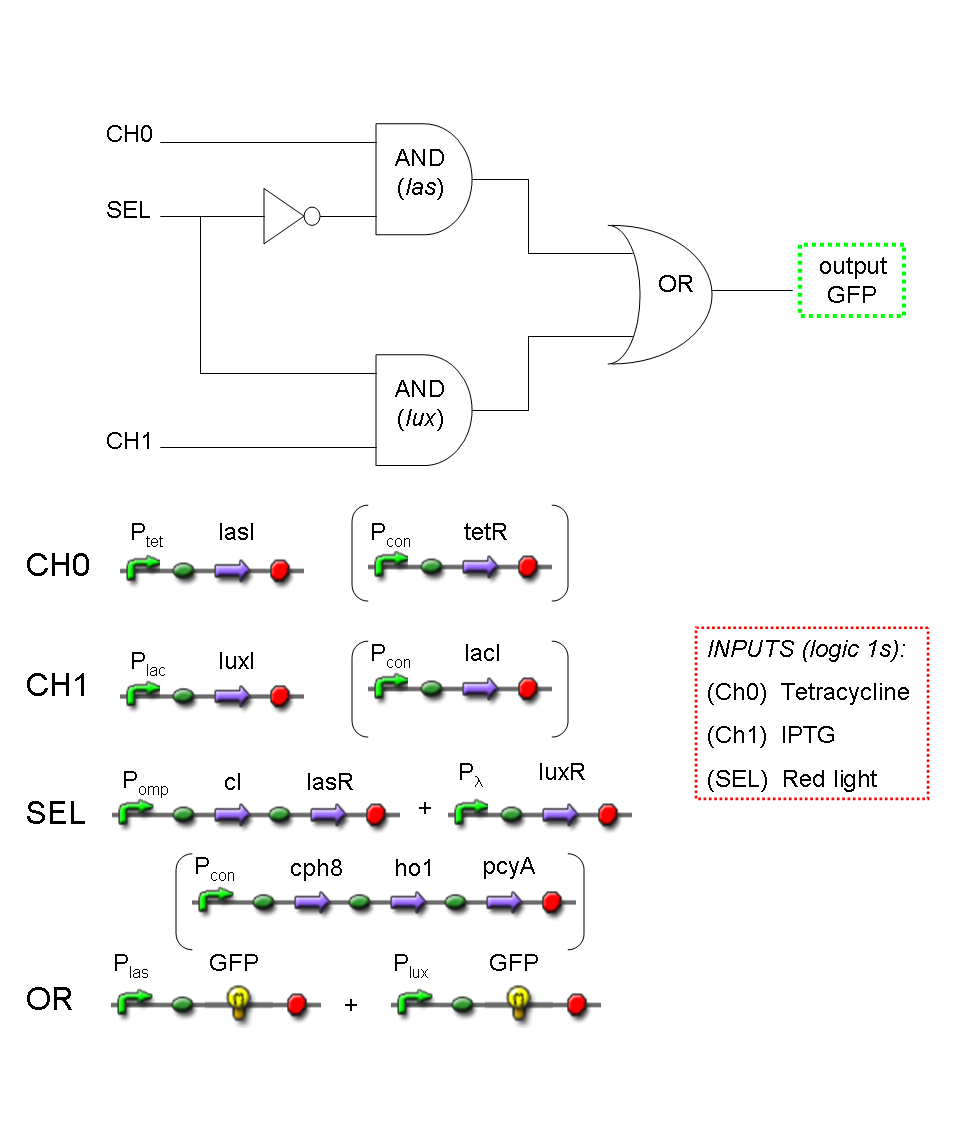
Mux and Demux logic circuits are composed by three fundamental logic gates, AND, OR, NOT: in the next paragraphs genetic implementation of these logic gates will be provided.
AND
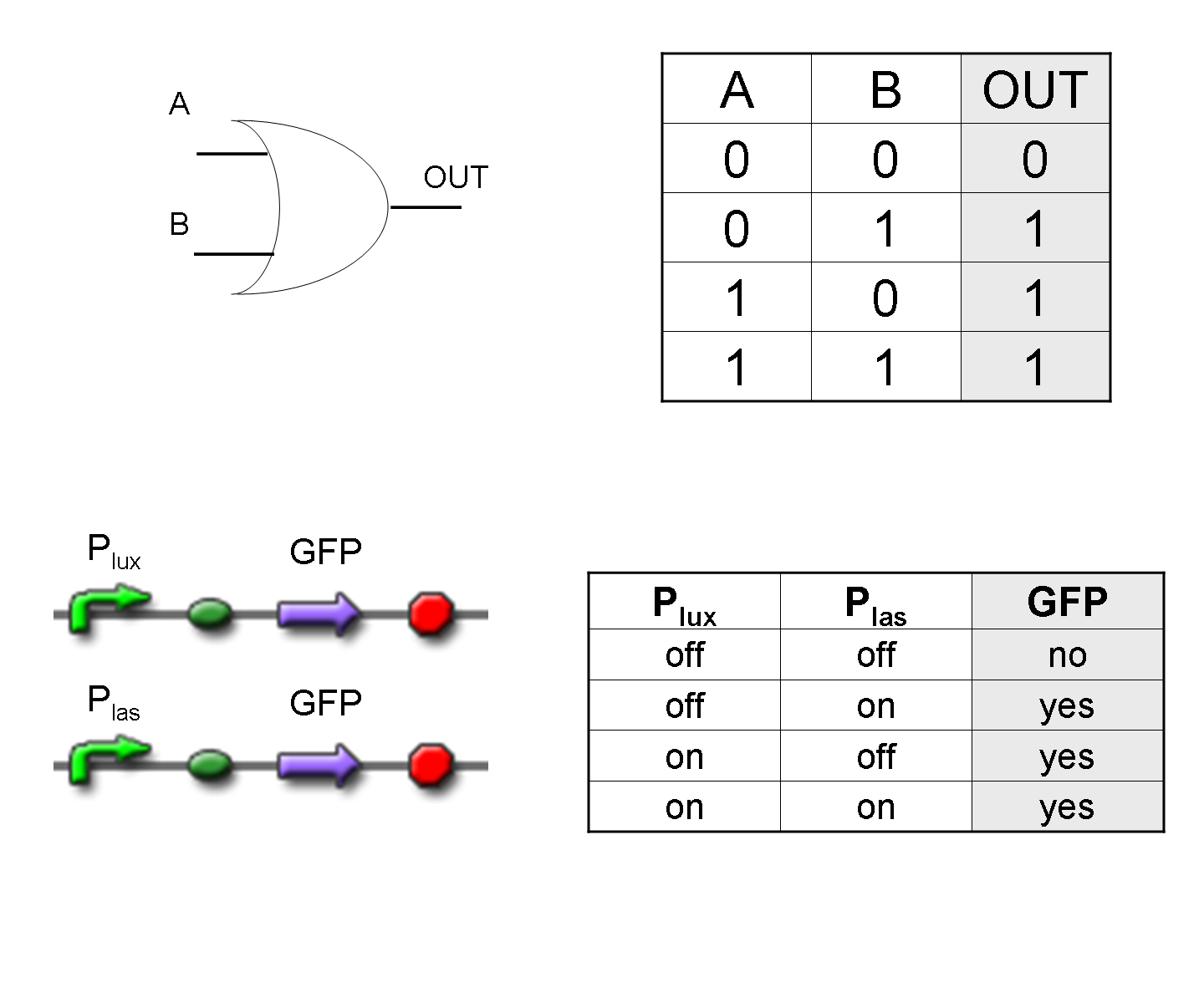
| To mimic an AND gate, we need a biological function, such as a promoter activation, which is directly turned on by the interaction between two upstream genes. In our synthetic devices, we use the luxR/luxI system: luxR can activate Plux promoter only upon 3-oxo-hexanoyl-homoserine lactone (HSL) binding; luxI generates HSL; so, only the contemporary expression of LuxR and luxI proteins can activate the downstream Plux-dependent gene expression. Another AND gate we use is the lasR/lasI system, which works in a very similar way but through another chemical intermediate, N-(3-oxododecanoyl) homoserine lactone (PAI-1). |
OR
| To mimic an OR gate in Mux, we need a biological function which can be activated alternatively by two independent upstream signals or by both. Thus, we combine the outputs of the upstream AND gates to assemble directly an OR reporter function, by simply repeating the reporter gene (GFP) under two different promoters (Plux and Plas). It’s sufficient to activate one of the two promoters (or both) to recover the GFP signal from engineered bacteria.
There should not be an over-expression problem for GFP, in fact, in Mux device, only one promoter can be active, either Plux or Plas. Here we considered GFP output, but OR device can be generalized for every output gene. |
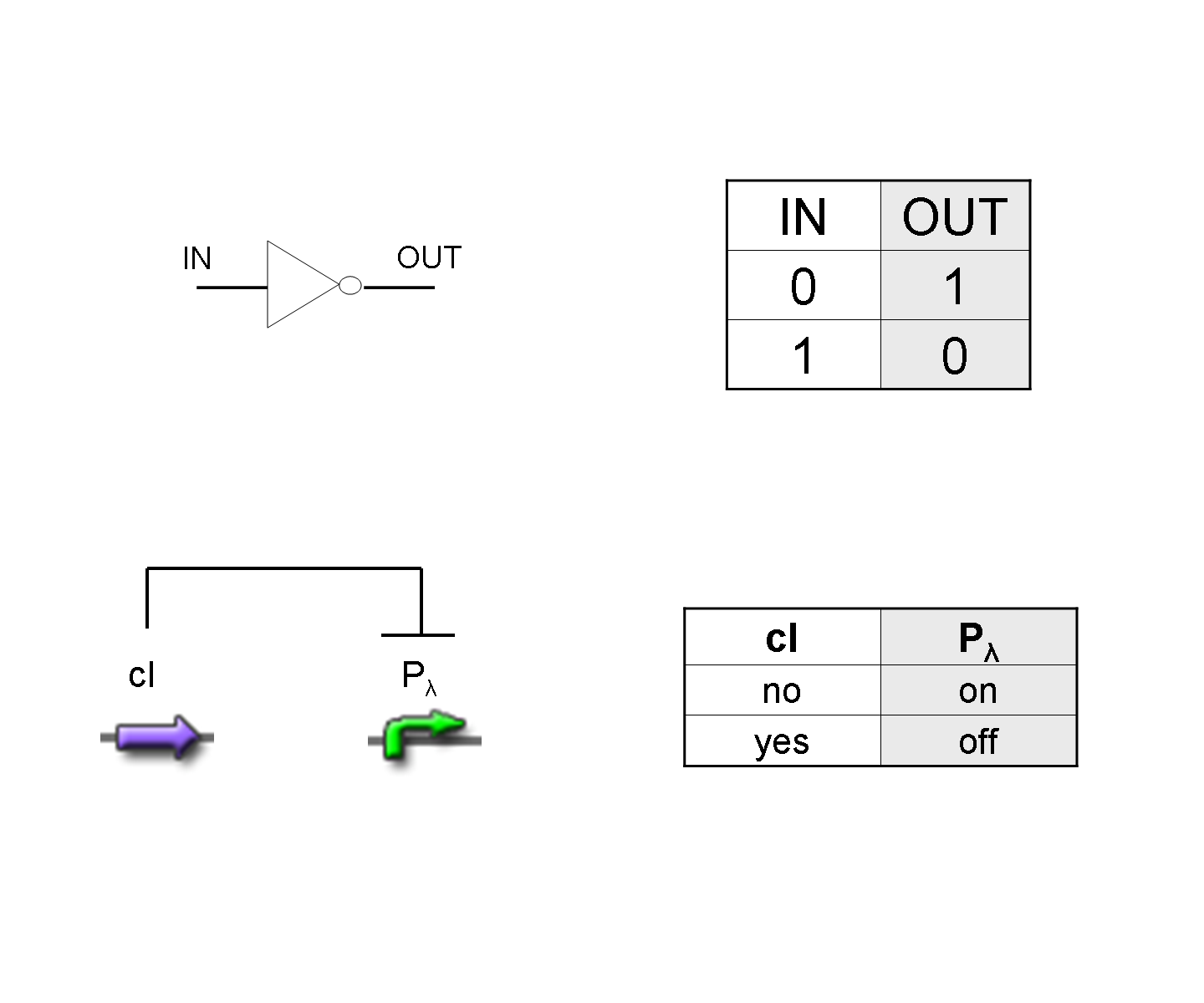
NOT
| To mimic a NOT gate, we need an efficient and regulated repressor of a specific downstream promoter: in this case, we choose cI repression on Plambda, which should be specific and, upon cI inactivation, quick and efficient. |
According to what above stated, genetic implementation of Mux and Demux can be obtained connecting these basic logic gates and can be summarized in this way:
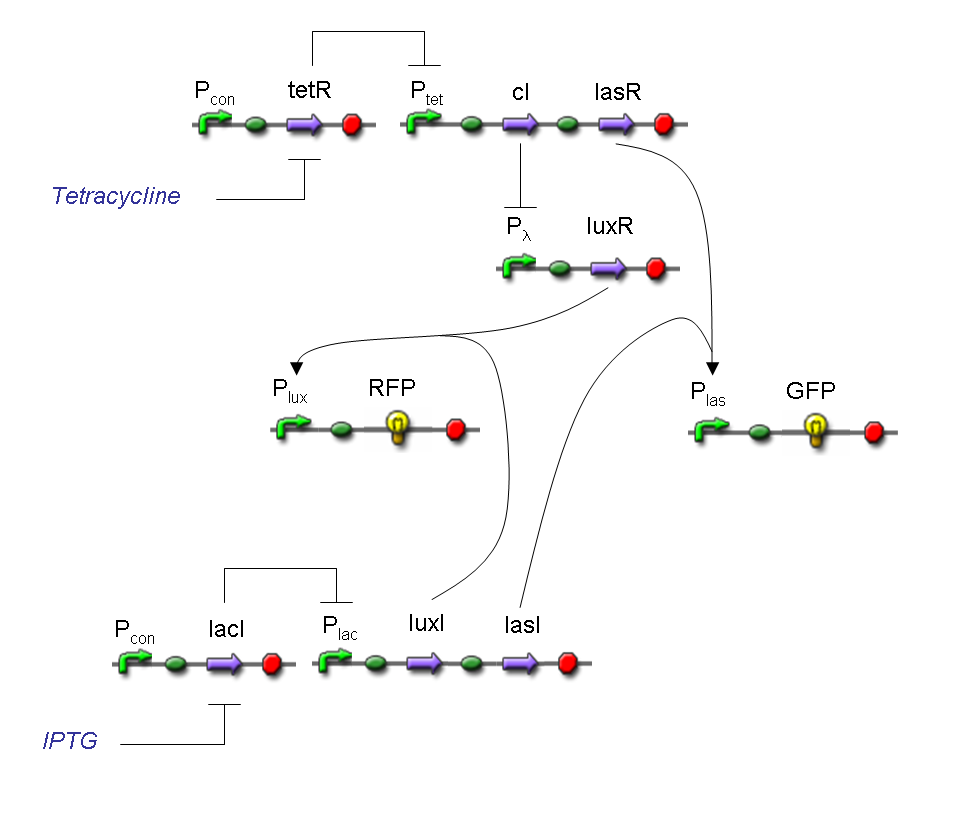
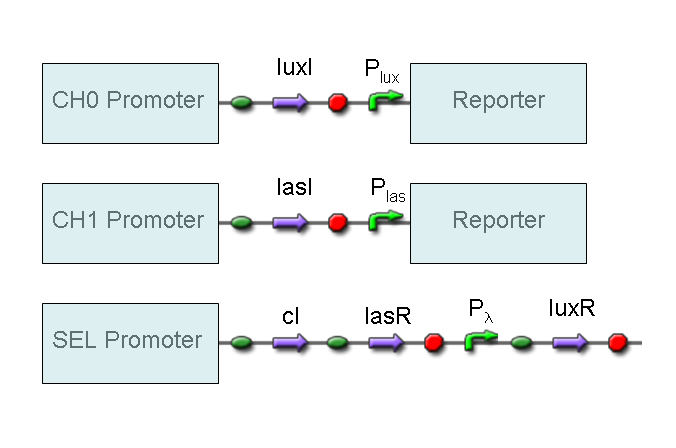
Genetic Mux
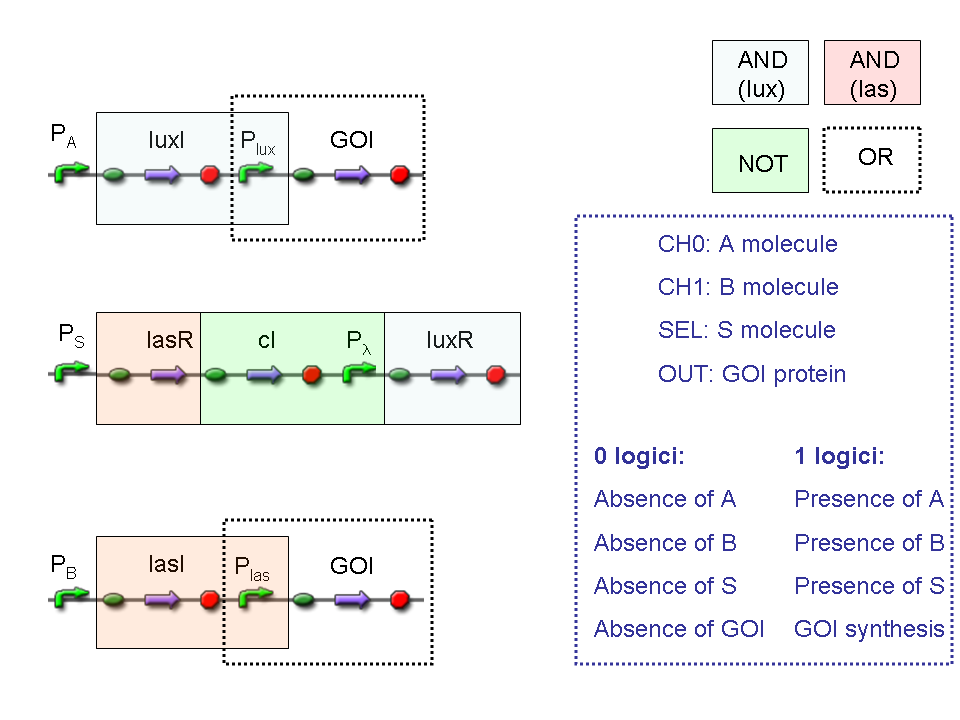
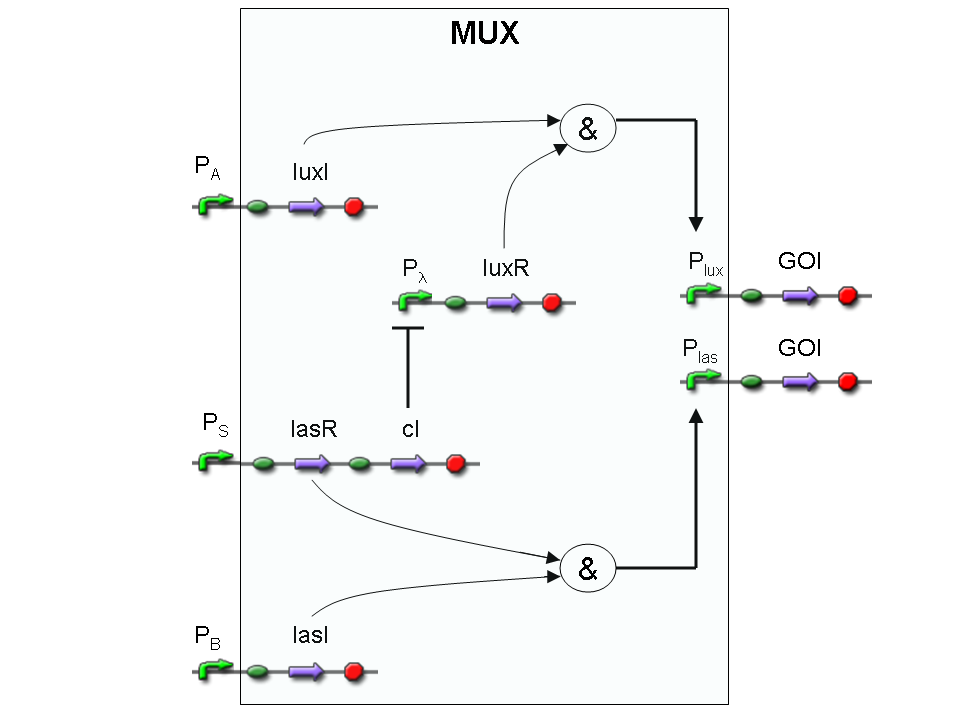
Let PA, PB and PS be three generic promoters that can be ACTIVATED respectively by the three exogenous molecules A, B and S. A genetic Mux with inputs A (CH0) and B (CH1), selector S and the generic protein GOI as output, can be implemented by the following gene network:
<\div>
We want to supply a device that can be generalized to detect every kind of input and to express every kind of gene as output. So, input promoters and output protein generators are not part of genetic Mux we want to build up. In this way, a hypotetical user can re-use our device assembling the desired input and output elements.
Now we report two examples of genetic Mux behavior, in response to generic inputs.
Genetic Mux behavior - Example 1
According to Mux truth table, if SEL=0, CH0=1, CH1=0, output channel must be logic 1. Red light is not present, so it can't dephosphorylate cph8-ho1-pcyA complex, which is constitutively expressed. cph8-ho1-pcyA complex activates Pomp promoter, so cI and lasR genes are transcripted. cI protein binds Plambda promoter and so luxR expression is inhibited. TetR is a repressor for Ptet promoter, but Tetracycline is present, so it can bind tetR protein, which is constitutively expressed, and can activate transcription of downstream gene: lasI. Simultaneous expression of lasI and lasR can activate GFP, which is under Plas promoter. IPTG is not present, so lacI protein binds Plac promoter and represses luxI transcription.
Genetic Mux behavior - Example 2
According to Mux truth table, if SEL=0, CH0=0, CH1=1, output channel must be logic 0.
Red light is not present, so it can't dephosphorylate cph8-ho1-pcyA complex, which is constitutively expressed.
cph8-ho1-pcyA complex activates Pomp promoter, so cI and lasR genes are transcripted.
cI protein binds Plambda promoter and so luxR expression is inhibited.
Tetracycline is not present, so tetR protein always binds Ptet promoter and lasI gene is not expressed.
There is no simultaneous expression of lasI and lasR, so Plas promoter can't be activated.
lacI is a repressor for Plac promoter, but IPTG is present, so it can bind lacI, which is constitutively expressed, and can activate transcription of downstream gene: luxI.
There is no simultaneous expression of luxI and lasI, so Plux promoter can't be activated.
These two examples show how red light can select the input to convey into the single output channel: in fact its presence allows lasR expression and represses luxR expression, while red light absence represses lasR expression and allows luxR expression.
Genetic Demux
We want to build up a device in which Input and Selector are respectively IPTG and Tetracycline.
In Demux we have two output channels: red fluorescence corresponds to logic 1 at Channel 0, while green fluorescence corresponds to logic 1 at Channel 1.
Absence of reporters expression corresponds to logic 0 at Channel 0 and Channel 1.
There isn't any input combination that corresponds to logic 1 at Channel 0 and Channel 1 together.
Genetic Demux behavior - Example 1
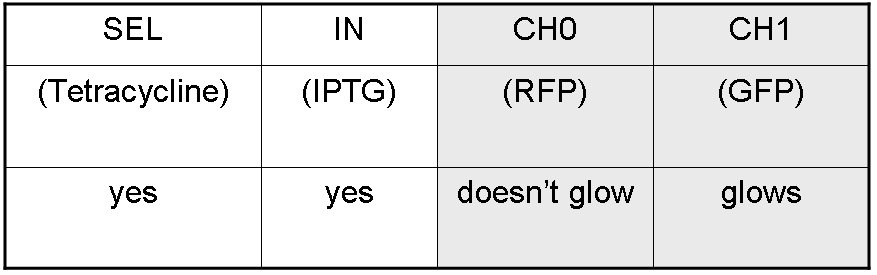
According to Demux truth table, if IN=1 and SEL=1 output channel 0 is logic 0 and output channel 1 is logic 1.
IPTG is present, so Plac promoter is active because IPTG binds lacI protein. This allows lasI and luxI transcription.
Tetracycline is present, so Ptet promoter is active because Tetracycline binds tetR protein. This allows cI and lasR transcription.
cI protein binds Plambda promoter and so luxR expression is inhibited.
Simultaneous expression of lasI and lasR can activate GFP, which is under Plas promoter.
There is no simultaneous expression of luxI and luxR, so RFP (which is under Plux regulation) cannot be expressed.
Genetic Demux behavior - Example 2
According to Demux truth table, if IN=0 and SEL=1 both output channels are logic 0.
IPTG is not present, so lasI and luxI are not expressed. This inhibits both Plux and Plas activation, because they behave like a logic AND in which one of the inputs is always 0.
Tetracycline presence is negligible because it allows lasR expression, but Plas cannot be active without lasI.
Example of a complete genetic Mux
Let PA, PB and PS be three generic promoters that can be ACTIVATED respectively by the three exogenous molecules A, B and S. A genetic Mux with inputs A (CH0) and B (CH1), selector S and the generic protein GOI as output, can be implemented by the following gene network:
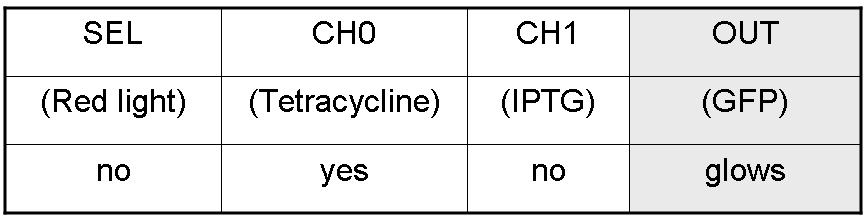
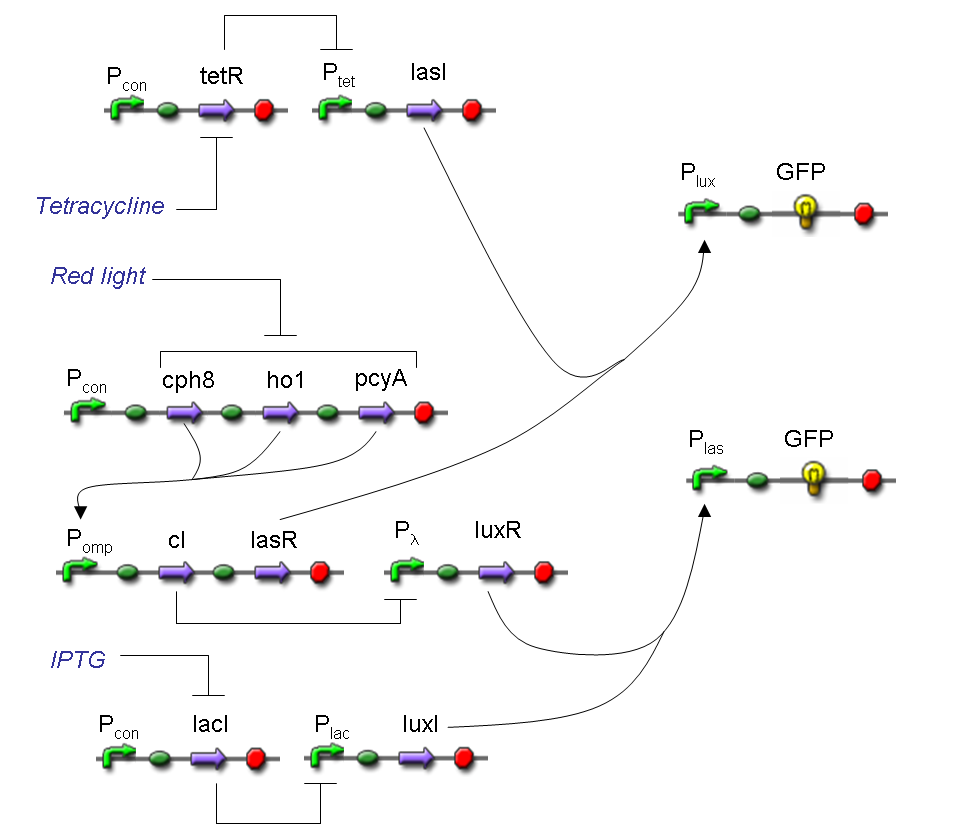
We want to build up a device in which Channel 0, Channel 1 and Selector are respectively sensitive to Tetracycline, IPTG and red light. The presence of each input corresponds to logic 1. We chose green fluorescence as Mux output: expression of GFP corresponds to logic 1, while absence of fluorescence corresponds to logic 0.
Complete genetic Mux - Example
According to Mux truth table, if SEL=0, CH0=1, CH1=0, output channel must be logic 1. Red light is not present, so it can't dephosphorylate cph8-ho1-pcyA complex, which is constitutively expressed. cph8-ho1-pcyA complex activates Pomp promoter, so cI and lasR genes are transcripted. cI protein binds Plambda promoter and so luxR expression is inhibited. TetR is a repressor for Ptet promoter, but Tetracycline is present, so it can bind tetR protein, which is constitutively expressed, and can activate transcription of downstream gene: lasI. Simultaneous expression of lasI and lasR can activate GFP, which is under Plas promoter. IPTG is not present, so lacI protein binds Plac promoter and represses luxI transcription.
Example of a complete genetic Demux
We want to build up a device in which Input and Selector are respectively IPTG and Tetracycline.
In Demux we have two output channels: red fluorescence corresponds to logic 1 at Channel 0, while green fluorescence corresponds to logic 1 at Channel 1.
Absence of reporters expression corresponds to logic 0 at Channel 0 and Channel 1.
There isn't any input combination that corresponds to logic 1 at Channel 0 and Channel 1 together.
Complete genetic Demux - Example
According to Demux truth table, if IN=1 and SEL=1 output channel 0 is logic 0 and output channel 1 is logic 1.
IPTG is present, so Plac promoter is active because IPTG binds lacI protein. This allows lasI and luxI transcription.
Tetracycline is present, so Ptet promoter is active because Tetracycline binds tetR protein. This allows cI and lasR transcription.
cI protein binds Plambda promoter and so luxR expression is inhibited.
Simultaneous expression of lasI and lasR can activate GFP, which is under Plas promoter.
There is no simultaneous expression of luxI and luxR, so RFP (which is under Plux regulation) cannot be expressed.
Applications
Genetic Mux and Demux have been described by now as systems which are able to detect fixed inputs (IPTG, Tetracycline and red light) and to return a fixed fluorescence (GFP and RFP) as output.
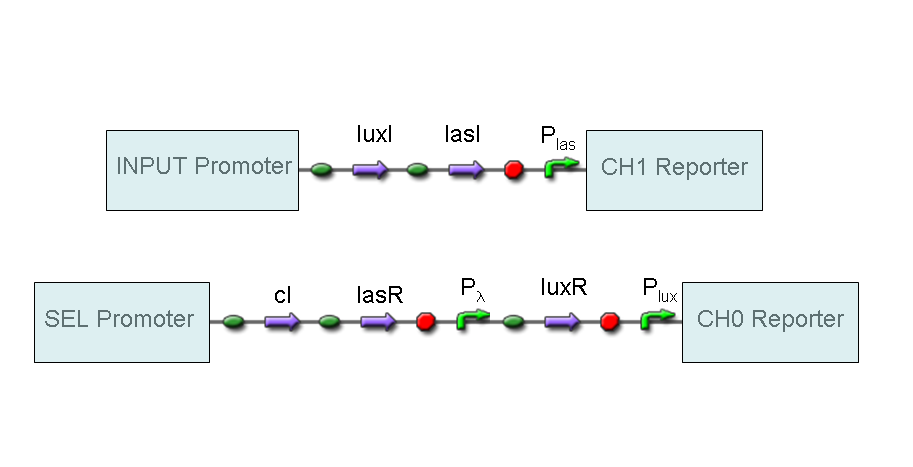
However, our purpose is to supply two devices (Mux and Demux) that can be generalized to detect every kind of input and to return every kind of reporter. To reach this goal, we set up a BioBrick assemble program in which parts corresponding to final devices can be re-used for any kind of input or output.
A hypothetical user of Mux or Demux only has to ligate our standard parts with desired promoters and reporters, as shown in the pictures above.
(In previous paragraphs, Channel 0 and Channel 1 in Mux have been cross-exchanged because red light input inhibits Pomp promoter and so Selector boolean signal is inverted).
 "
"