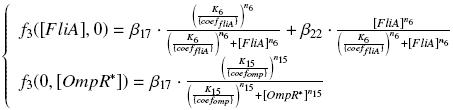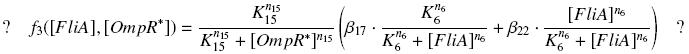Team:Paris/Modeling/f3
From 2008.igem.org
We have [OmpR*] = {coefomp}expr(pTet) = {coefomp} ƒ1([aTc]i)
and [FliA] = {coefFliA}expr(pBad) = {coefFliA} ƒ2([arab]i)
So, at steady-states,
| param | signification | unit | value | comments |
| [expr(pFlhDC)] | expression rate of pFlhDC with RBS E0032 | nM.min-1 | need for 20 mesures with well choosen values of [aTc]i and for 20 mesures with well choosen values of [arab]i and 5x5 measures for the relation below? | |
| γGFP | dilution-degradation rate of GFP(mut3b) | min-1 | 0.0198 | |
| [GFP] | GFP concentration at steady-state | nM | need for 20 + 20 measures and 5x5 measures for the relation below? | |
| (fluorescence) | value of the observed fluorescence | au | need for 20 + 20 measures and 5x5 measures for the relation below? | |
| conversion | conversion ratio between fluorescence and concentration | nM.au-1 | (1/79.429) |
| param | signification corresponding parameters in the equations | unit | value | comments |
| β13 | production rate of FliA-pFlhDC with RBS E0032 β13 | nM.min-1 | ||
| (K12/{coeffliA}) | activation constant of FliA-pFlhDC K12 | nM | ||
| n12 | complexation order of FliA-pFlhDC n12 | no dimension | ||
| β2 | production rate of OmpR-pFlhDC with RBS E0032 β2 | nM.min-1 | ||
| (K22/{coefomp}) | activation constant of OmpR-pFlhDC K22 | nM | ||
| n22 | complexation order of OmpR-pFlhDC n22 | no dimension |
Then, if we have time, we want to verify the expected relation
 "
"