From 2008.igem.org
[http://2008.igem.org/Team:The_University_of_Alberta  ]
[http://2008.igem.org/Team:The_University_of_Alberta/Team
]
[http://2008.igem.org/Team:The_University_of_Alberta/Team  ]
[http://2008.igem.org/Team:The_University_of_Alberta/Project
]
[http://2008.igem.org/Team:The_University_of_Alberta/Project  ]
[http://2008.igem.org/Team:The_University_of_Alberta/Parts
]
[http://2008.igem.org/Team:The_University_of_Alberta/Parts  ]
[http://2008.igem.org/Team:The_University_of_Alberta/Notebook
]
[http://2008.igem.org/Team:The_University_of_Alberta/Notebook  ]
]
"How do I love thee, plastic? Let me count the ways. I wake up and glance at my plastic digital cable box to check the time. I go to the bathroom to use my plastic toothbrush, shaking a bit of my “nontoxic” tooth powder from a plastic bottle. I fill the plastic container of my Waterpik with mouthwash from another plastic bottle. I step into the shower—my lacy white curtain is protected by a plastic liner, and my chlorine-free shower water comes to me through a plastic-encased filter.Ah, but in the kitchen I am a bit freer of you, plastic. When I learned that my plastic bowls, dishes, and containers could leach harmful chemicals—especially the ones with that sneaky, practically invisible little recycling triangle embossed with the number 7—I bought Pyrex. My soft-boiled eggs are served up in a Pyrex glass dish. A moment of rebellion against thee, plastic! But the microwave in which I heat water for tea is made of plastic as well as metal. And the refrigerator shelf on which I store my eggs is plastic.
The coaster on my desk, on which I place my steaming, oh-so-healthy green tea, is plastic. The 22-inch liquid-crystal computer monitor that seems to be the fulcrum of my entire existence is made of plastic. My keyboard, my mouse, my computer speakers, the CD cases for my music collection, my polycarbonate reading glasses, the remote control for my stereo, my telephone—all plastic. The sun shines through my window onto a riot of green plants in … plastic pots. (I could switch to ceramic, but it’s so heavy and hard to heft when I want to water them.) And I’ve been up for only an hour."
[http://discovermagazine.com/2008/may/18-think-you-can-live-without-plastic/(by Jill Neimark in Discovery Magazine 04.18.2008]
See also the article that inspired our project at [http://discovermagazine.com/2008/may/18-the-dirty-truth-about-plastic | Link]
The Project
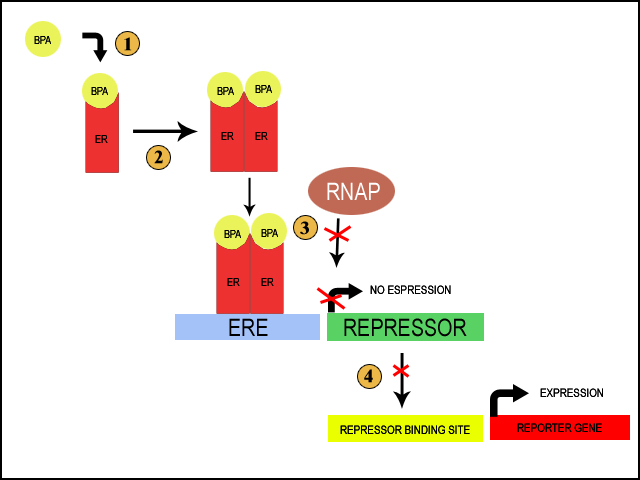
Estrogen Receptor Mechanism
The aim of our project is to create a biosensor that will detect BPA and other estrogenic compounds in the environment. BPA is a toxin that has been shown to leech from certain types of plastic. Studies have shown this chemical to have detremental effects in animal studies and is very likely to be harmful to humans as well. Having a biosensor that can detect soil and water contaminated with BPA would be a useful tool indeed.
Our project relies on the mechanism by which BPA affects cells. BPA is an estrogenic compound; i.e. it has the ability to mimic estrogen. When taken into the cell, it can bind to the cytoplasmic estrogen receptors, and activate genes related to estrogen, tricking cells into thinking they are in an environment containing estrogen when they are not.
Our biosensor will take advantage of the estrogen receptor (see diagram to the right)
We aim to move the human estrogen receptor (hER) into E.Coli, which will activate a reporter system in the presence of BPA. When BPA is present in the environment, it difuses through the cell membrane and binds to free cytoplasmic estrogen receptor (1). This causes the estrogen receptors to dimerize (2). Dimerization of the estrogen receptors allows for the receptors to beind to the estrogen responsive element (ERE); this is a small sequence of DNA that the receptors can specifically bind once they are dimerized (3). The ERE is placed immeadetly upstream of the promotor for a repressor. The binding of the reseptor should then block the binding of RNA Polymerase and prevent the transcription of the repressor. Under normal circumstances, the repressor would itself prevent the transcription of a reporter gene (Purple Russian/Blue Ox). Thus, when BPA is in the environment, the repressor is repressed and the reporter gene is actively transcribed, giving us a quick visual way to determine the presence of BPA (4).
Next, plan on intorducing BPA degrading genes from Sphingomonas bisphenolicum to aid in the remediation of BPA contamination. Ultimately, we aim to put this biosensor/degradation system into plants - More on this later.
Info
- bisphenol degrading gene from the Sphingomonas bisphenolicum strain AO1
[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nuccore&id=171363657 Link]
Status
At this point our operon is nearly complete. All bricks downstream of the estrogen receptor have been assembled, with only the estrogen receptor to be his-tagged and given a promotor before we add the rest onto the end. We have acquired BPA for testing.
Journal Articles of Interest
Synthetic Receptors
An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants Jianru Zuo, Wi-Wen Niu and Nam-Hai Chua
[http://www.blackwell-synergy.com/doi/full/10.1046/j.1365-313x.2000.00868.x?prevSearch=allfield%3A%28chua%29| Link]
Protein Complementation
A ToxR-based two-hybrid system for the detection of periplasmic and cytoplasmic protein–protein interactions in Escherichia coli: minimal requirements for specific DNA binding and transcriptional activation Frank Hennecke, Arne Müller, Roland Meister, Astrid Strelowand Susanne Behrens1,6
[http://peds.oxfordjournals.org/cgi/content/full/18/10/477| Link]
A -Galactosidase-Based Bacterial Two-Hybrid System To Assess Protein-Protein Interactions in the Correct Cellular Environment Jimmy Borloo, Lina De Smet, Bjorn Vergauwen, Jozef J. Van Beeumen, and Bart Devreese
[http://pubs.acs.org/cgi-bin/abstract.cgi/jprobs/2007/6/i07/abs/pr070037j.html| Link]
Estrogen Receptors
Estrogen response element-dependent regulation of transcriptional activation of estrogen receptors and ß by coactivators and corepressors C M Klinge, S C Jernigan, K A Mattingly, K E Risinger and J Zhang
[http://jme.endocrinology-journals.org/cgi/content/full/33/2/387| Link]
Cooperative Coactivation of Estrogen Receptor in ZR-75 Human Breast Cancer Cells by SNURF and TATA-binding Protein Bradley Saville, Hetti Poukka, Mark Wormke, Olli A. Jänne, Jorma J. Palvimo, Matthew Stoner¶, Ismael Samudio, and Stephen Safe
[http://www.jbc.org/cgi/content/full/277/4/2485| Link]
Identification of a Novel RING Finger Protein as a Coregulator in Steroid Receptor-Mediated Gene Transcription Anu-Maarit Moilanen, Hetti Poukka, Ulla Karvonen, Marika Häkli, Olli A. Jänne, and Jorma J. Palvimo
[http://mcb.asm.org/cgi/content/full/18/9/5128| Link]
High level expression of full-length estrogen receptor in Escherichia coli is facilitated by the uncoupler of oxidative phosphorylation, CCCP Zhang C.C.; Glenn K.A.; Kuntz M.A.; Shapiro D.J.1
[http://www.ingentaconnect.com/content/els/09600760/2000/00000074/00000004/art00120;jsessionid=22tnudxvlkl5k.alice?format=print| Link]
Nuclear receptor coactivators function in estrogen receptor- and progestin receptor-dependent aspects of sexual behavior in female mice Heather A. Molenda-Figueiraa, Casey A. Williamsa, Andreana L. Griffina, Eric M. Rutledgeb, Jeffrey D. Blausteina and Marc J. Tetela
[http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WGC-4K5HWP0-2&_user=1067472&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000051251&_version=1&_urlVersion=0&_userid=1067472&md5=e1dd22c068b2b8288136f34eab6b6514| Link]
Estrogen receptor interaction with estrogen response elements Carolyn M. Klinge
[http://nar.oxfordjournals.org/cgi/content/full/29/14/2905#SEC2| Link]
A novel estrogen sensor based on recombinant Arxula adeninivorans cells: Thomas Hahna, Kristina Taga, Klaus Riedela, Steffen Uhligb, Keith Baronianc, Gerd Gellissend and Gotthard Kunzea
[http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6TFC-4J32JC8-1&_user=1067472&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000051251&_version=1&_urlVersion=0&_userid=1067472&md5=2e20cf8bb0b465392aa852b10cda184b| Link]
Estrogen receptor ß: an overview and update: Chunyan Zhao, Karin Dahlman-Wright, and Jan-Åke Gustafsson
[http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2254331 Link]
A simple and extremely sensitive system for detecting estrogenic activity using transgenic Arabidopsis thaliana
[http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WDM-4G314HG-2&_user=1067472&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000051251&_version=1&_urlVersion=0&_userid=1067472&md5=1b0120c5a09915ebdf83160641791af0| Link]
How estrogen-specific proteins discriminate estrogens from androgens: a common steroid binding site architecture
[http://www.fasebj.org/cgi/content/full/17/10/1334 Link]
Bisphenol A
Direct Evidence Revealing Structural Elements Essential for the High Binding Ability of Bisphenol A to Human Estrogen-Related Receptor-γ: Hiroyuki Okada, Takatoshi Tokunaga, Xiaohui Liu, Sayaka Takayanagi, Ayami Matsushima, and Yasuyuki Shimohigashi
[http://www.ehponline.org/members/2007/10587/10587.html| Link]
Endocrine disruptor bisphenol A strongly binds to human estrogen-related receptor γ (ERRγ) with high constitutive activity:Sayaka Takayanagia, Takatoshi Tokunagaa, Xiaohui Liua, Hiroyuki Okadaa, Ayami Matsushimaa and Yasuyuki Shimohigashi
[http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6TCR-4KT6GTR-3&_user=1067472&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000051251&_version=1&_urlVersion=0&_userid=1067472&md5=f58023e52189a0ee9ef7fd6263dff43f| Link]
Aerobic degradation of bisphenol A by Achromobacter xylosoxidans strain B-16 isolated from compost leachate of municipal solid waste:Chang Zhanga, Guangming Zenga, Li Yuana, Jian Yub, Jianbing Lia, Guohe Huanga, Beidou Xie and Hongliang Liua.
[http://www.sciencedirect.com.login.ezproxy.library.ualberta.ca/science?_ob=ArticleURL&_udi=B6V74-4N0XNBH-2&_user=1067472&_coverDate=05%2F31%2F2007&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000051251&_version=1&_urlVersion=0&_userid=1067472&md5=60133aec998135b6157590f860fb5d3d| Link]
Molecular cloning and characterization of cytochrome P450 and ferredoxin genes involved in bisphenol A degradation in Sphingomonas bisphenolicum strain AO1: M. Sasaki, T. Tsuchido and Y. Matsumura.
[http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2672.2008.03843.x Link]
Processing of Bisphenol A by Plant Tissues: Glucosylation by Cultured BY-2 Cells and Glucosylation/Translocation by Plants of Nicotiana tabacum: Nobuyoshi Nakajima1,4, Yukiko Ohshima2, Shigeko Serizawa2, Tomoko Kouda2, John S. Edmonds2, Fujio Shiraishi2, Mitsuko Aono3, Akihiro Kubo3, Masanori Tamaoki1, Hikaru Saji3 and Masatoshi Morita2.
[http://pcp.oxfordjournals.org/cgi/content/full/43/9/1036 Link]
Purification of Cytochrome P450 and Ferredoxin, Involved in Bisphenonol A Degradation, from Sphingomonas sp. Strain AO1: Miho Sasaki,Ayako Akahira, Ko-ichi Oshiman, Tetsuaki Tsuchido, and Yoshinobu Matsumura.
[http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1317416 Link]
Biodegradation of bisphenol A by cells and cell lysate from Sphingomonas sp. strain AO1: Miho Sasaki1, Jun-ichi Maki1, Ko-ichi Oshiman3, Yoshinobu Matsumura1, 2 and Tetsuaki Tsuchido1, 2.
[http://www.springerlink.com/content/q7864l02734wg32m/fulltext.pdf Link]
Bisphenol A Degradation by Bacteria Isolated from River Water: J.-H. Kang and F. Kondo.
[http://www.springerlink.com/content/ge1mec9289r4cw3w/fulltext.pdf Link]
|
 "
"
 ]
] ]
] ]
] ]
] ]
]