A. Genetic Engineering and Part Construction
Saccharomyces cerevisiae are widely used for baking and brewing, and they are particular useful for synthesizing metabolites under fermentation conditions which prevent the air oxidation of many useful compounds. To achieve our project and expand the synthetic biology toolbox for programming yeast, we have introduced into the iGem registry BioBricks encoding 3 yeast promoters, 3 yeast terminators, a two micron origin of replication, 2 selectable markers, 2 enzymes, and a yeast integration plasmid. In addition, we have generated seven constructs using these parts. Furthermore, we have submitted two additional parts representing a foundational tool, including a gene encoding an amber suppressed RFP biobrick for screening of SupF+ (Amber suppressor) genotype and an amber suppressor tRNA biobrick.
Yeast Regulator Biobricks
Three unique constitutive and repressible yeast promoters were introduced into the registry.
Yeast Promoters
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K122000 BBa_K122000] |
pPGK1 |
This is the 1500 bp upstream of the PGK1 coding region in an industrial yeast strain. Constitutive promoter. |
1497 |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K122002 BBa_K122002] |
pADH1 |
700bp upstream of ADH1 promoter region containing RBS. Constitutive promoter. |
701 |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K122001 BBa_K122017] |
pGAL1 + tetO |
The GAL1 promoter which is highly repressed by glucose. An additional tetracycline operator site was included upstream of the RBS to allow repression by tetR. |
484 |
Four additional yeast terminators were introduced into the registry.
Yeast Terminators
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K122003 BBa_K122003] |
tCYC1 |
300bp downstream the CYC1 coding region in a standard yeast strain. |
300 |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K122004 BBa_K122004] |
tADH1 |
300bp downstream the ADH1 coding region in a standard yeast strain. |
300 |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K122013 BBa_K122013] |
tPGK1 |
1000bp downstream the PGK1 coding region in an industrial yeast strain. |
1000 |
Selectable Markers
Selectable Markers
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K122001 BBa_K122018] |
ZeoR |
Zeocin Resistance Gene |
300 |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K122008 BBa_K122008] |
BleoR |
Bleocin Resistance Gene under pTet promoter |
800ish |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K122014 BBa_K122014] |
ORI+HisTag |
2 Micron ORI and Histadine Tag |
<9000 |
Project Specific Constructs
This year's project involved the integration of 4-coumarate:coenzyme A ligase/ stilbene synthase fusion protein, a tyrosine ammonia lyase, and a zeocin resistance selectable marker. For more information, please visit Strategy.
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K122001 BBa_K122001] |
[pGAL1][tetO][ZeoR] |
 |
|
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K122005 BBa_K122005] |
Tyrosine Ammonia Lyase |
 |
|
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K122010 BBa_K122010] |
4CL:STS |
 |
|
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K122012 BBa_K122012] |
[pPGK1][4CL:STS][tCYC1] |
 |
|
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K122015 BBa_K122015] |
[pGAL1][tetO][ZeoR][tADH1] |
 |
|
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K122021 BBa_K122021] |
[pADH1][TAL][tPGK1] |
 |
|
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K122016 BBa_K122019] |
[pPGK1][4CL:STS][tCYC1][pGAL1][tetO][ZeoR][tADH1] |
 |
|
Additional Bacterial Parts
Novel Zero Leak Inverter Concept
wtRFP in SupF- / ARFP in SupF- /wtRFP in SupF+ / ARFP in SupF+
Please visit our Notebook for a summary of labwork, protocols, and overviews of progress.
B. Yeast Transformation
C. HPLC Data
To analyze our beer samples for resveratrol content, be will be using High Performance Liquid Chromatography (HPLC), which will allow us to separate the metabolites produced by the yeast and analyze these compounds by spectrophotometry. By comparing HPLC chromatogram peaks of metabolites produced by our yeast with a resveratrol-only standard, we can identify if resveratrol is being produced, and at what quantities. Below, we show our initial data for HPLC calibration curves using known quantities of resveratrol and p-coumaric acid standards and test chromatograms using extracts from different wine samples.
- HPLC Parameters
- Column: Agilent Eclipse XDB-C18, 5uM (9.4x250mm)
- Mobile Phases:
- (A) 5% acetonitrile / 0.95% acetic acid
- (B) 70% acetonitrile / 0.3% acetic acid
- Linear gradient: A to B over 29 minutes
- Flow rate: 0.9mL/min
- Absorbance monitoring: 290nm
- Sample injection volume: 25 microliters
|
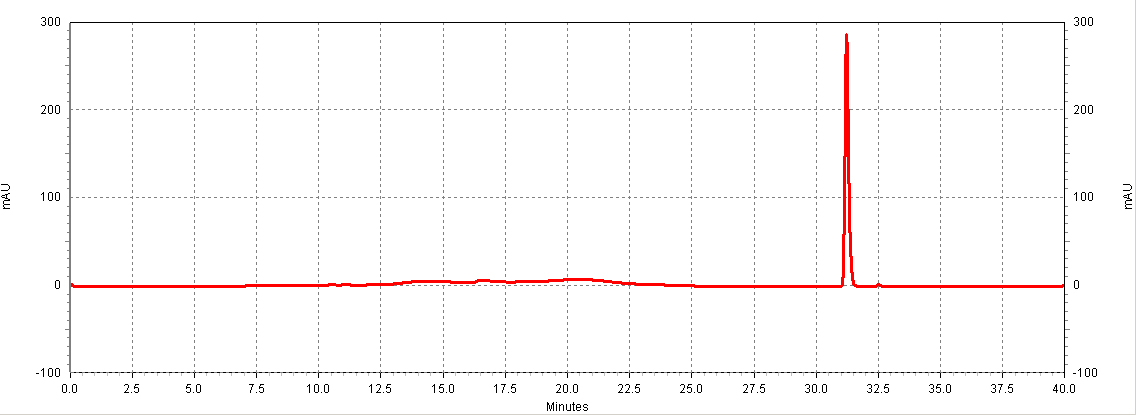 Figure C1. Example HPLC chromatogram of a resveratrol standard. HPLC: Calibration
HPLC: Fermenation batches
Fermentation
Coming Soon. For a sneak preview, check out the Gallery
|