Team:Newcastle University/ProofOfConceptBrick
From 2008.igem.org
Newcastle University
GOLD MEDAL WINNER 2008
| Home | Team | Original Aims | Software | Modelling | Proof of Concept Brick | Wet Lab | Conclusions |
|---|
Home >> Proof of Concept Brick
The designs for regulatory networks produced by our software are too large and complicated to be implemented in the time available. We needed a smaller system to try out our ideas.
We wanted to show that it was possible to build a system to sense quorum peptides. Bacillus subtilis ATCC 6633 produces a small peptide antibiotic, subtilin, that is also used in quorum sensing. We decided to develop a system that would sense subtilin and incorporate it into B. subtilis 168, a strain that does not naturally possess this peptide sensing system.
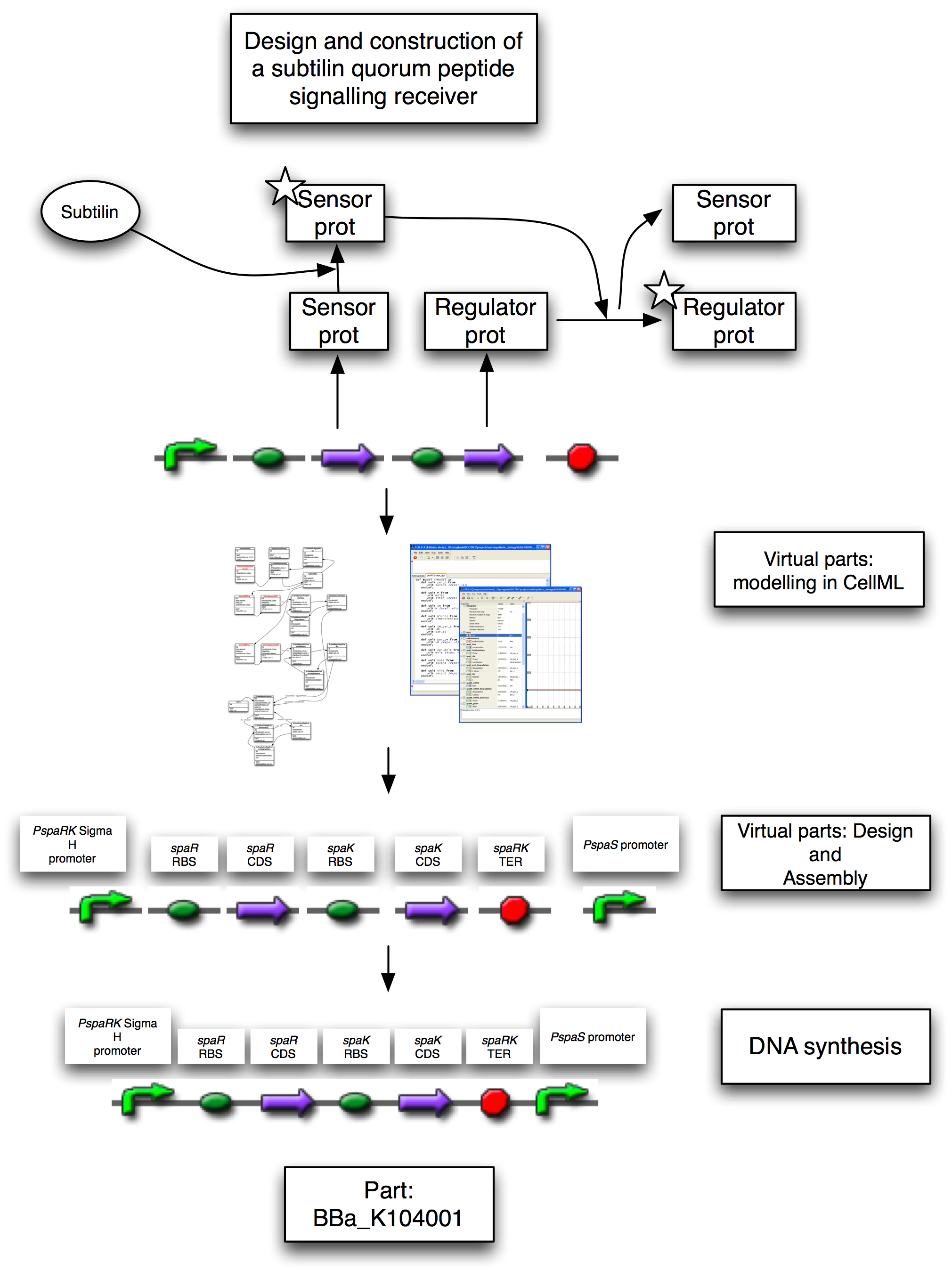
To demonstrate that the sensory and regulatory systems will work in isolation from the genes for subtilin production, we generated a proof-of-concept brick. This contains the two-component system that senses extracellular subtilin and in response activates genes with response elements in their promoters. The brick also contains one such promoter, at the far right-hand-side. The rational was that by simply placing this brick directly up-stream of any promoterless gene, that gene can be induced by the presence of extracellular subtilin, allowing us to rapidly produce positive controls for the two-component system.
In the original strains, a single operon contains all of the genes responsible for subtilin production and export, sensing, and down-stream gene regulation. For the BugBuster application, we needed to be able to pick up subtilin secreted by the unmodified strain, within our modified bacterium. This meant that our bacterium needed the genes for sensing subtilin, and for activating genes in response to it, but must not themselves produce subtilin. We re-designed the system in a bottom up fashion from virtual parts encoded in CellML.
Figure 1: Design and implementation of the subtilin receiver BioBrick, [http://partsregistry.org/Part:BBa_K104001 BBa_K104001]. This contains a promoter, driving a reduced spa operon, separated from a PspaS promoter by a terminator. The reduced spa operon contains the CDS for spaR and spaK only, with spaK being the subtilin sensor which activates spaR, the transcription factor. The PspaS promoter is activated by the spaR transcription factor. On sensing the subtilin signal peptide, the spaK subtilin sensor activates the spaR regulatory protein by phosphorylation. This activate form now activates the transcription of other genes. In the full system, these would be other sensors or transcription factors. In the proof-of-principle positive controls, these would be reporters, such as GFP that respond to the increase in POPs.
References
C Klein, C Kaletta, N Schnell and K D Entian (1992). Analysis of genes involved in biosynthesis of the lantibiotic subtilin. Appl Environ Microbiol. 58(1): 132-142.
C Klein, C Kaletta and K D Entian (1993). Biosynthesis of the lantibiotic subtilin is regulated by a histidine kinase/response regulator system. Appl Environ Microbiol. 59(1): 296-303.
M. Kleerebezem, R. Bongers, G. Rutten, M. W. de Vos and O. P. Kuipers. (2004). Autoregulation of subtilin biosynthesis in Bacillus subtilis: the role of the spa-box in subtilin-responsive promoters. Peptides. 25(4): 1415-1424
 "
"