Team:Hawaii/Initial Synth. Oligo Assembly
From 2008.igem.org
Contents |
Protocol
Hybridization of Parts
- Mix:
- 3 μl 100 µM sense oligo
- 3 μl 100 µM anti-sense oligo
- 3 μl 10 x PNK (polynucleotide kinase) buffer
- 2 μl 10mM ATP
- 2 μl T4 polynucleotide kinase (PNK)
- 17 μl distilled water
- Total volume = 30 μl
- Incubate at 37C for 1.5 hours.
- Add 4 μl 0.5 M NaCl.
- Place in boiling water bath for 2 min., then remove water bath from the heat source and allow the reaction (still in the water bath) to cool to room temperature (approx. 30 minutes)
- Reference: Pam Silver Lab. [http://openwetware.org/wiki/Silver:_Oligonucleotide_Inserts| Oligonucleotide Inserts.]
DNA Ligation
- Combine 4 ul of construct one with 4 ul of construct 2.
- Add 1 ul of nanopure H20.
- Add 10 ul of 2x quick ligation reaction Buffer. In order to avoid shearing the DNA, mix by resuspending very slowly.
- Add 1 ul of Quick T4 DNA ligase and mix thoroughly by resuspending very slowly.
- Incubate at room temperature for 5 minutes.
- Cool on ice then transform, or store at -20oC
- Reference: Quick Ligation Kit from NEB.
Restriction Enzyme Digest
- May not be necessary
- Add REs in appropriate amounts to appropriate buffer (check if enzymes are compatible).
- Incubate at least 1 hour at recommended temperature (usually 37C).
- Stop reaction by adding 5 μl 10X loading buffer or by incubating 10 min. at 65C.
Notes:
- <10% of digest should be RE
- REs are stored in glycerol. <5% glycerol should be present in digest. Therefore, a minimum 10-fold dilution is necessary so the enzymes don't act funky.
- It's always a good idea to gently shake and spin down RE and RE buffer prior to use.
- Reference: Short Protocols in Molecular Biology. Vol. 1. 5th edition. 3-3 to 3-4.
Results
First attempt
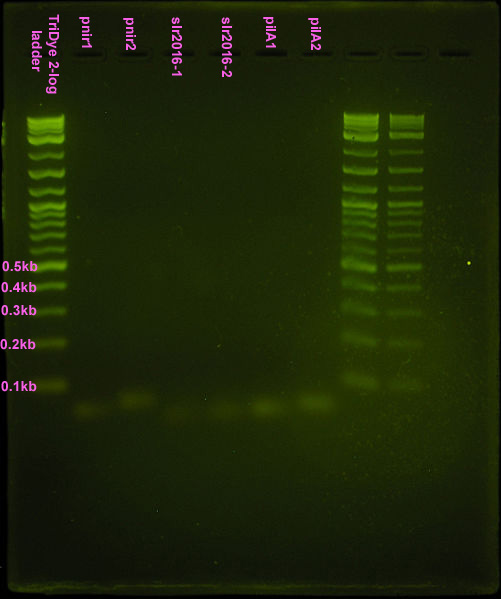
Nice bands observed for annealed oligos. Bands appear to be the correct size (<100bp) but we can't tell if anneal worked (EtBr doesn't tell us if these are ssDNA or dsDNA bands). Smear is probably from loading way too much DNA (see discussion). Possible faint band for pnir ligation. No bands for slr2016 and pilA ligation, probably because way too much DNA was added for the ligation reaction.
Second attempt
Did a ligation using 1 μl of a 10-6 dilution of each annealed product. Ligated at both room temperature and at 40C (recommended by Frank, etal). Nothing was visible on the EtBr stained 4% agarose gel because not enough DNA was present.
Third attempt
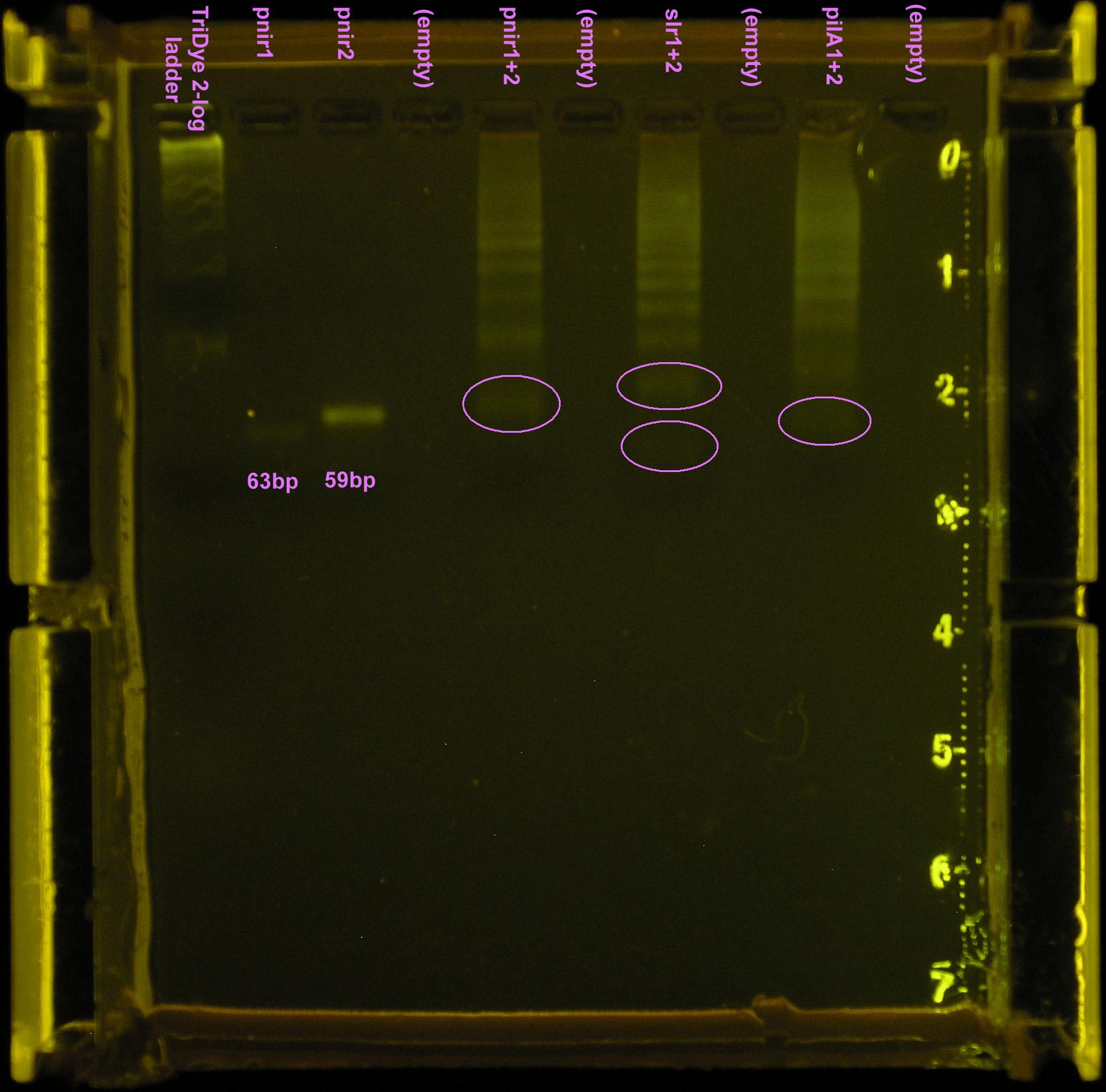
Did a ligation using 10-2 dilutions of annealed product (ligated at room temperature and at 40C). Faint bands were visible for the annealed products (10-2 dilutions) but no bands were observed for the ligated products. Not enough DNA was present. Ran 10-1 dilutions of annealed product on SYBR Safe stained 2% agarose gel.
Fourth attempt
Took 10 μl ligation reaction from first attempt and restriction digested with XbaI and PstI (1 μl each) in NEBuffer 3 (1 μl). A ladder of bands were observed for the ligation product, indicating incomplete digestion.
Fifth attmpt
Discussion
We need to dilute our annealed products before running them in a gel or attempting ligation. 20-125ng DNA (per lane) is good for running gels. One reaction using the NEB Quick Ligation Kit can ligate up to 180ng of DNA.
 "
"