Team:Caltech/Project/Phage Pathogen Defense
From 2008.igem.org
|
People
|
Phage Pathogen Defense
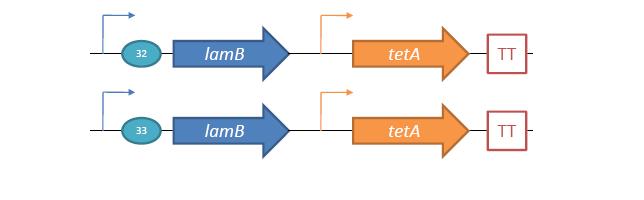
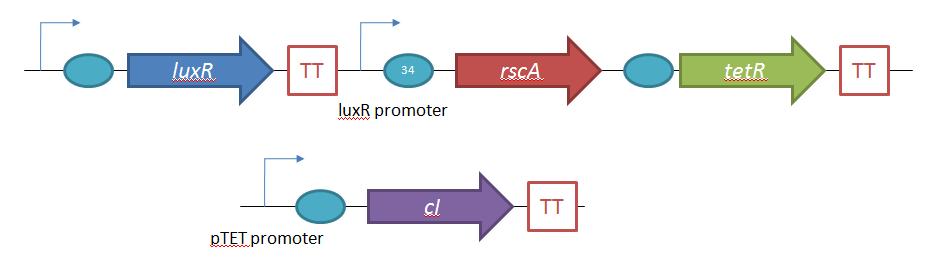
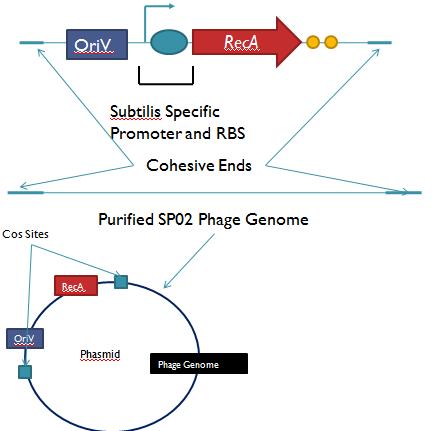
General IdeaBacterial food poisoning is a prevalent problem around the world; in the year of 2006, the United States had more than 325,000 hospitalizations from food borne illnesses. The iGEM 2008 team is designing a probiotic for medical applications, and given the prevalence of foodborne illnesses, pathogen defense is high on the list of priorities. Bacteriophages were chosen because they have evolved to be highly effective at infecting and killing their bacterial hosts, and their highly infectious and replicative nature means just one phage can lead to decimation of the pathogen population. Two methods have been used to approach the goal of ‘manufacturing’ phage. The first utilizes the Escherichia coli bacteriophage λ as a defense against pathogenic E. coli. A strain of E. coli has been developed which is immune to λ phage infection, but is lysogenic for λ. Furthermore, a plasmid has been developed to control the induction of λ using the E. Coli gene rscA, thus, with some application of signal, the lysogens can be triggered to enter the lytic cycle and release phage into the environment to infect other E. Coli. Due to the fact that the probiotic is immune to λ phage infection, the λ phage will only target pathogenic E. coli. The second method developed can be adapted to any temperate bacteriophage to target many species of bacteria. By circularizing phage DNA with a E. Coli plasmid origin of replication, the phage resides within the probiotic as a plasmid, however, upon conjugation with a bacteria of the host species, the ‘phasmid’ will produce virulent phage to destroy the pathogenic host population. Part I: Lambda PhageBacteriophage λ is a temperate phage with an E. Coli. host, λ infects E. Coli through the lamB receptor, and absence of this receptor prevents λ infection. Our project takes advantage of this aspect of bacteriophage λ to create E. Coli which are resistant to the phage, but release the phage to destroy susceptible pathogenic E. Coli. To achieve this, lamB deficient E. Coli must first express the surface protein through a constitutively active version of the gene on a plasmid. This allows the lamB deficient E. Coli to be infected by λ phage. Lysogens are selected for using antibiotic resistance, and then the plasmid possessing the lamB receptor gene is counter-selected against, producing a strain lysogenic for λ, but is immune to infection. System DesignThe system design revolves around the construction of two plasmids, both of which to be placed within E. Coli Strain JW3996-1, a strain deficient in the maltose outer membrane porin lamB, a surface protein integral to λ phage infection. One of these two plasmids is first responsible for creating JW3996-1 λ lysogens. However, creating lamB- lysogens is complicated by the fact that the JW3996-1 strain is immune to λ phage infection. Thus, to allow for λ infection, lamB must be expressed on a plasmid. The gene coding for lamB was obtained from E. Coli genomic DNA using PCR. For regulation of lamB expression, a weak constitutive promoter, J23113, and 2 weak ribosomal binding sites, B0032 and B0033, were placed upstream of the lamB gene. In the final system, the lysogens will have to be immune to infection, thus, the lamB+ plasmid will have to be cured from the JW3996-1 strain. This is proposed to be done via fusaric acid tetracycline counter-selection [4], a procedure which will allow for selective pressure against tetracycline resistance. To apply this to the lamB+ plasmid, a tetracycline resistant cassette (P1005) with a terminator (B0015) has been cloned downstream of the lamB gene. The second plasmid which must be designed controls the induction of the lysogens to release phage into the environment. Control of this aspect of the system is vital for integration with the overall iGEM project. In general, λ lysogens stay in lysogeny until some trigger, usually cellular stress. However, we want to be able to induce the lysogens into the lytic cycle, this is done through the over expression of the E. Coli gene rscA, which has been shown to bring lysogens into the lytic phase. Currently, the rscA gene has been placed behind the luxR repressor/activator. This repressor prevents expression of rscA until activation via acyl-homo-serine lactone (AHL). However, within the final project, rscA will most likely be placed in the control of Allen's random differentiation generator. Furthermore, the final system will be similar in nature to Doug's, with a inverter after the activation of rscA, this is shown below. Current WorkWill be updating when it is not 6 am. ConstructsSame as above Part II: B. Subtilis LysogensBasic IdeaWe wish to create a phasmid, basically the lysogen genome with an E. Coli plasmid origin of replication, using B. Subtilis lysogens. This allows the phage genome to pass on as a plasmid within our engineered E. Coli, but when the plasmid is conjugated to B. Subtilis, the virus is induced and destroys the pathogens. Phasmid construction will an E Coli origin of replication with a Subtilis specific promoter in front of RecA or another inducer. Current ProgressThree different bacteriophage lysogens and three wildtype strains of B. Subtilis has been ordered from the Bacillus Stock Center. However, efforts to induce the bacteriophage from the lysogens with UV exposure has proved to be futile thus far, however, this was possibly due to a contamination of the lysogen stocks. The strains have been reordered and B. Subtilis lysogen induction with UV occurring soon, most likely the week of 8/4. |
 "
"