Home
People
Project Details
Protocols
Completed Systems
Biosafety
Support
|
Subprojects
Note: Click on the subproject title or picture for a detailed description of the subproject
|
|
Folate is the generic term for the various forms of Vitamin B9, which include dihydrofolate (DHF), tetrahydrofolate (THF), and folic acid. An essential vitamin for cell survival, folate is involved in amino synthesis (and thus DNA synthesis) as well as single-carbon-transfer reactions. Though humans don't produce folate, folate deficiency can cause serious birth defects and anemia. As a result, most cereals and breads are supplemented with folate. Folate has also been shown to have good bioavailability in the large intestine, and so is more easily absorbed by the host than other vitamins.
Our aim is to engineer a strain of E. coli that will overexpress folate such that a person without access to green, leafy vegetables or folate-supplemented foods can still obtain the necessary daily amount by having this strain residing in their gut.
|
|
Approximately 75% of adults worldwide suffer from lactose intolerance, the inability to metabolize lactose in the small intestine. We propose to treat lactose intolerance by engineering a strain of Escherichia coli that can reside in the large intestine. The engineered strain will sense lactose and subsequently release ß-galactosidase to convert lactose into glucose and galactose, both of which can be reabsorbed by the host. To treat lactose intolerance, our engineered bacterial strain will contain two plasmids: one with constitutive expression of a mutant lactose permease and ß-galactosidase, and the second with lactose-inducible expression of the lambda phage lysis cassette. The mutant lactose permease allows the cells to import lactose under all conditions. When the cells uptake enough lactose, the second plasmid will induce cell lysis through activation of the lambda phage lysis cassette, resulting in cell lysis and release of ß-galactosidase into the large intestine. Data covering the construction and characterization of these plasmid constructs is discussed if you click the link above :-).
|
|
|
|
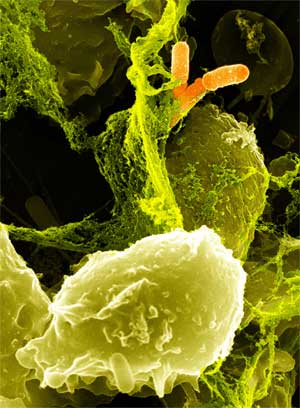
Several types of bacteria can cause illness in humans by infecting the gut. Salmonella and E. coli are probably the two people most frequently associate with food poisoning. There are other pathogenic bacteria that can infect our gut as well. Shigella and Campylobacter can cause cramping, diarrhea and dysentery. Vibrio cholerae, which also infects the gut, is the cause of cholera. The body normally relies on white blood cells (neutrophils) to clear bacteria from the body. Once a bacterium is engulfed, the neutrophil releases a sudden and toxic amount of reactive oxygen species, comprised of a mixture of superoxide, hydrogen peroxide, and hypochlorous acid. This is event is termed the oxidative burst. However, white blood cells do not patrol the gut lumen and so there is no active clearance of pathogens. The goal of this project is to engineer a beneficial gut microbe capable of detecting a harmful bacterium and, in turn, generate an oxidative burst sufficient to kill the pathogen.
|
|
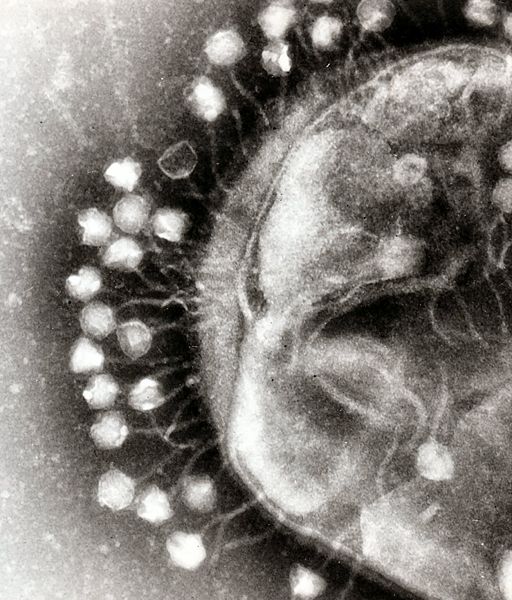
Another aspect of bacterial pathogen defense for our probiotic is to produce bacteriophages, which would rapidly infect and wipe out all of the pathogens. There are basically methods to approach phage production, differentiated by the type of phage used. The first uses the bacteriophage λ, which targets E. Coli. The other is exploring the use of a temperate bacteriophage from B. Subtilis, however this method, if successful, can be adapted to temperate bacteriophages of any bacterial strain.
Bacteriophage λ is a temperate phage with an E. Coli. host, λ infects E. Coli through the lamB receptor, and absence of this receptor prevents λ infection. We will takes advantage of this aspect of bacteriophage λ to create E. Coli which are resistant to the phage, but release the phage to destroy susceptible pathogenic E. Coli.
The second approach is more versatile, and can target more strains of pathogenic bacteria. The goal is to create a phasmid out of the genome of a temperate bacteriophage. A phasmid combines a E. Coli plasmid Origin of Replication with a linear phage genome, circularizing it. This allows the phasmid to pass on as a plasmid within E. Coli, however when transferred to its native host, the phage phage is induced.
|
|
|
|
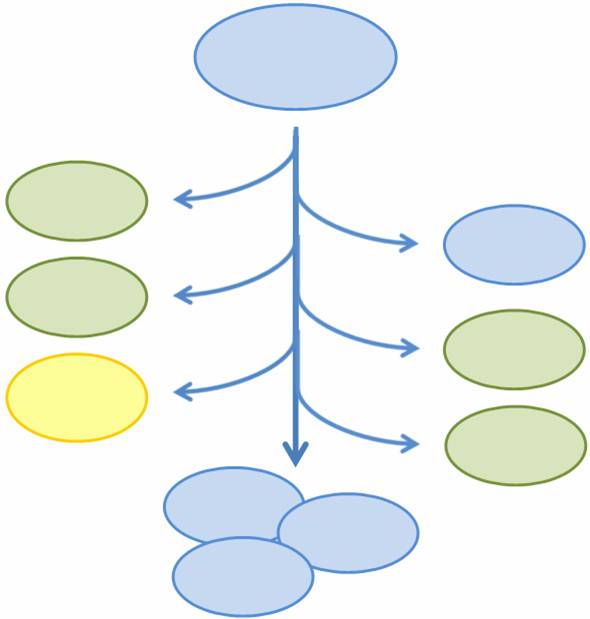
Given that we have created four different states for a cell to be in, we need to in some manner combine them into one system. However, we need to ensure that any particular cell is in only one of these states, and not more than one, or else the load on the cell may be too big. In other words, we want to make these states mutually exclusive.
To do so, two new devices have been created: a randomly activated off-to-on switch and a population variation generator. Initially, a cell is in a default state (S0), the switch is off, and the fate of the cell is undetermined. However, each time a cell replicates the plasmid that contains the switch, there is a chance that the switch turns on. Once the switch is on, it activates the population variation generator, which in turn determines the fate of the cell by setting it to one of three states - S1, S2, or S0 (the original default). That cell and all of its descents then stay in the determined state.
|
|
 "
"