Team:Caltech/Project/Population Variation
From 2008.igem.org
|
People
|
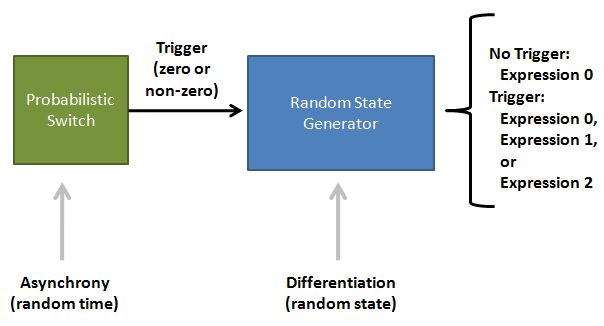
Population Variation: Construction of an Asynchronous Random State Generator
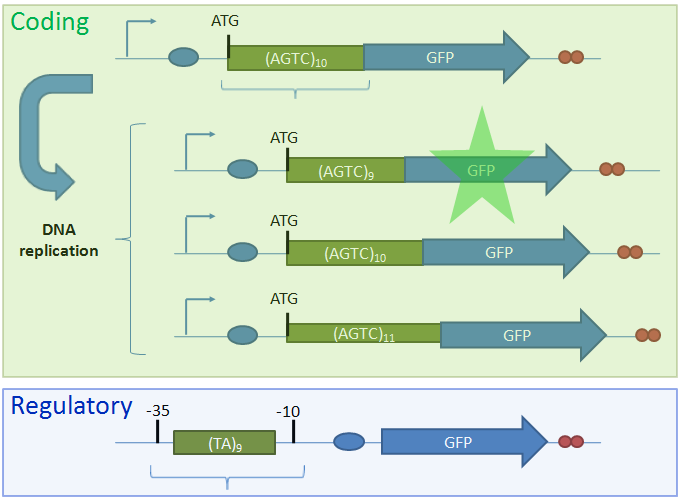
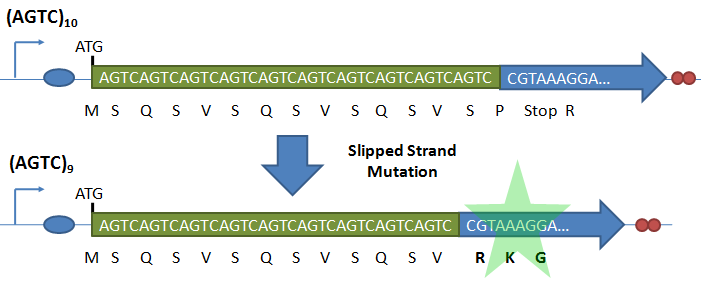
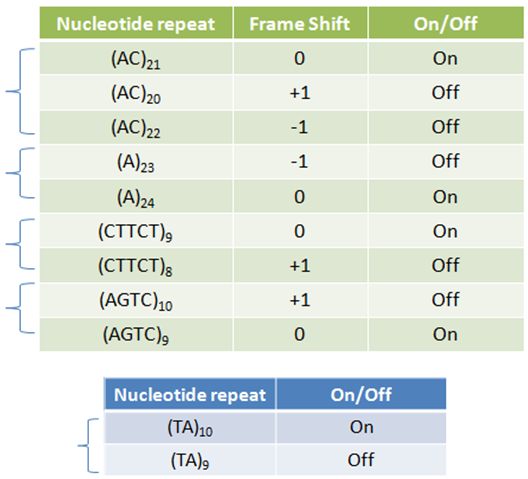
IntroductionBiological circuits are commonly introduced into cells to produce non-native functions. Such functions include the production of antimalarial drugs in yeast cells1, the detection of light by bacterial cells2, and the enzymatic breakdown of bacterial biofilms by bacteriophages3. However, a problem arises when dealing with functions that inhibit the reproduction of cells. For example, the overproduction of certain compounds may burden cells with such a heavy load that they grow at a slower rate. Such production may involve lysing the cell to release compounds into solution. Once a cell is lysed, it can no longer produce progeny. If all the cells in a colony lyse, then the cell line may eventually die. One solution is to introduce cellular differentiation. Two states can be created in a cell: one in which the cell simply grows and replicates, and another in which the fatal function is turned on. Cells in the former state can produce progeny, and the progeny that they produce can either be in the former or latter states. Cells that differentiate into the latter state can die off without directly affecting the viability of the cell line. Thus, differentiation allows for the incorporation of fatal functions into cells. Random StateDifferentiation can also be used to combine multiple non-compatible functions into a single cell line. In this case, only one function will be turned on in each cell at any time, but the cell population will express the entire set of functions. Two functions may be non-compatible if they compete for the same resources inside a cell or if they signal through the same intracellular components. These two functions could be simply placed into separate cell lines, but if these two cell lines have different growth rates, one cell line would outcompete the other. Differentiation separates different functions into individual cells and allows the entire cell line to express multiple functions. In such a system, cells start off in an undifferentiated state. As undifferentiated cells replicate, their offspring can stay in the current state or switch into one of the mutually-exclusive states. As long as cells divide faster than they differentiate, the reproduction of undifferentiated cells maintains the undifferentiated population, which ensures that the entire cell line is kept alive. Thus, fatal and mutually-exclusive functions can be incorporated into cell lines by utilizing cellular differentiation. Random TimeIt is also important to have cells differentiate at random times. Having asynchrony ensures that not a large portion of the population will differentiate at once. Doing so might leave too few undifferentiated cells to maintain the population. It is hard to preserve the condition that cells divide faster than they differentiate if there are bursts in time in which many cells differentiate into terminal states. In addition, if the environment the cells are in continuously changes, cells may differentiate into a particular state that is incompatible with the current environment. For example, that particular function may be useless in the current environment, or the performing that function in such an environment may lead to the cell’s death. Thus, spreading out the time during which cells differentiate increases the chances that an individual cell committed to a state finds itself in a hospitable environment. Thus, both random state and random time are needed for differentiation. System OutlineTo implement cellular differentiation, we designed two BioBrickTM devices. The first device is a probabilistic switch that randomly activates the second device. The second device is a random state generator that, when triggered, determines the final state of the bacterial cell line. Combined, the two devices form a system that allows for the integration of multiple de novo functions into bacterial cell lines. Probabilistic Switch: Slipped Stand MutationsTo construct the first device, the probabilistic switch, we exploit the fact that DNA polymerase slips when replicating long stretches of short nucleotide repeats5,6. This phenomenon is called slipped-strand mutation (SSM). We have created two families of such switches based on where the long repeat is integrated. The first family, called coding SSMs, places a repeat in between the start codon of a gene and the remaining coding region. To create an on-to-off switch, the gene is initially placed in-frame with the start codon, and thus the gene is correctly translated and expressed. When DNA polymerase slips and miscopies the stretch of nucleotide repeats, the coding sequence shifts out of frame with respect to the start codon, and the gene is no longer correctly translated7,8. In an off-to-on switch, the coding sequence is initially out of frame and can either go into the correct frame or stay out of frame when SSM occurs, depending on whether the stretch of repeats becomes a multiple of three. The second family, called regulatory SSMs, places the repeat between the -10 and -35 elements of a promoter. To create an on-to-off switch, the distance between the -10 and -35 elements is initially optimal for sigma factor binding. The sigma factor helps recruit RNA polymerase to the promoter and initiate transcription. When the polymerase slips and changes the number of nucleotide repeats, the distance between the -10 and -35 elements changes. Suboptimal spacing between promoter elements reduces sigma factor recognition, resulting in less frequent transcription events8,9.
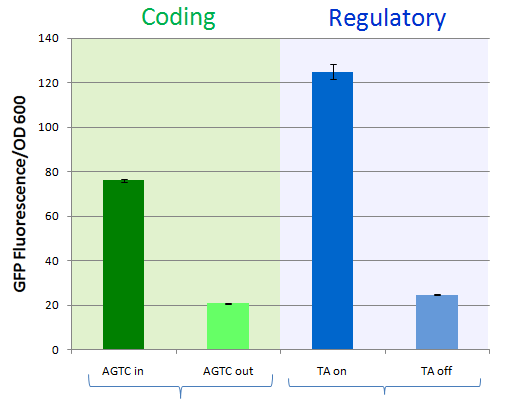
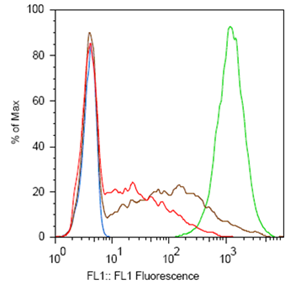
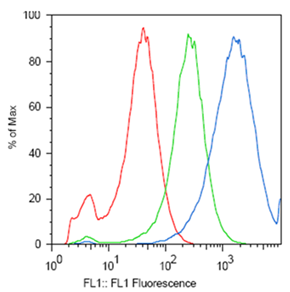
ResultsTo test the functionality of SSMs as genetic switches, we used the green fluorescent protein (GFP). By measuring the fluorescence of bacterial cells that contain our constructs, we could determine the expression of GFP in the on and off states. We constructed four classes of coding SSMs, and one class of regulatory SSMs. Each class contains one in-frame or ‘on’ variant, and one or more out-of-frame or ‘off’ variants. The fluorescence of these constructs was then measured for the population and at the single-cell level. We found that AGTC coding repeats and TA regulatory repeats display detectable GFP fluorescence in the ‘on’ state and significantly reduced fluorescence in the ‘off’ state. From flow cytometry data, a broad distribution in fluorescence was observed in cells with AGTC constructs covering three orders-of-magnitude. Since the constructs were placed in high copy plasmids, these distributions might reflect hundreds of individual GFP coding sequences shifting in and out of frame within each cell.
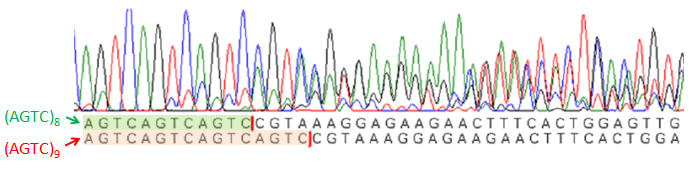 Sequencing results suggest slipped-strand mutation. Cells were transformed with just the coding SSM construct (AGTC)9. However, the sequencing trace for a colony transformed with (AGTC)9 also shows the presence of (AGTC)8, suggesting that slippage at the AGTC repeat had occurred. The sequence for GFP after the start codon follows the AGTC repeats. Future WorkIn future work, we would use the SSM as a switch to turn on the gene that codes the trigger for the second device. To implement a coding SSM, we would place repeats after the start codon of the trigger gene. The trigger would initially be off and turned on when the polymerase slips. Or, to implement a regulatory SSM, we would place the trigger gene downstream of a SSM promoter. Then the gene would be transcribed when the promoter slips into the right length to bind to a transcriptional activator. We would also place coding and regulatory SSMs in front of LacZ, and then chromosomally integrating the construct into E. coli so that there is only one copy of the construct in a cell. Our current work utilizes a high copy number plasmid, and it is difficult to resolve individual slipped strand mutations and calculate slippage frequency in such cases. With LacZ, blue-white screening can then be conducted to determine the switching frequency of the SSMs. In addition, we plan to place the SSMs in front of the gene that codes for tetracycline resistance (tetA). We can then select for and against cells with tetracycline resistance. Random State Generator: FimE Recombinase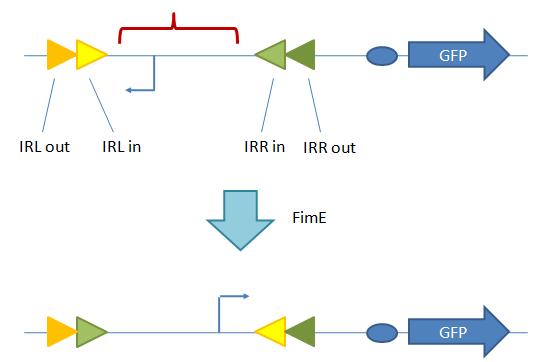 FimE flips the region flanked by IRL and IRL. A tetR promoter is placed between the two binding sites facing upstream. When the DNA segment is flipped, the promoter faces downstream, and GFP is produced. Once the in and out components of a binding site do not match, FimE no longer recognizes that site. The length of the recombination region (in red) was varied from 250 bp to 850 bp in increments of 100 or 200 bp. FimE is the trigger that will act on the random state generator. FimE is a recombinase protein that can flip DNA flanked by two binding sites. The random state generator consists of recombination regions flanked by FimE binding sites. FimE recognizes binding sites called IRR (inverted repeat right) and IRL (inverted repeat left). Each IRL and IRR consists of an inner and outer sequence, and fimE will only recognize a site that has matching inner and outer sequences. After FimE flips the DNA in between two correctly formed binding sites, the binding sites become distorted, and FimE can no longer act on the region. Taking advantage of this effect, one could place a promoter initially pointing upstream in between the two binding sites, and a gene downstream of the recombination region. Since the promoter is pointing in the opposite direction of the gene, the downstream gene will not be transcribed. However, once FimE flips the flanked recombination region, the promoter then points in the correct direction and can transcribe the downstream gene10. ResultsGiven that we were placing a different DNA segment in between the two binding sites than is found in wild-type genes, we wanted to determine the range of lengths FimE will act on and how the length affects FimE activity. In addition, we want to determine if the ratio of determined states could be skewed by changing the lengths of different recombination sites. Thus, we tested how the length of the recombination site affects FimE’s ability to detect the two flanking binding sites. The fluorescence of cells with each construct was used as an indicator of FimE activity. We observed a relationship between GFP fluorescence and separation length (Figure 7). Maximum fluorescence was observed to occur at about a separation length of 250 bp. This distance may be a function of entropy and enthalpy. The chances that two DNA segments on the same strand come together statistically decrease as the two segments are separated by intervening sequences. In addition, two sites very close together are unlikely to come into contact with each other, as it is energetically unfavorable to bend DNA to such a great degree. These two opposing forces might explain the presence of an optimal separation length. Future WorkUtilizing the fact that FimE flips DNA and cannot recognize binding sites that it has distorted, we can design a system that uses two IRR and two IRL sites to produce a total of three possible states, depending on how FimE acts. Once FimE flips any region in this system, it cannot flip the DNA segment again. Hence, the state of the cell is established after FimE acts. This assumes that FimE is unable to flip adjacent IRL and IRR sites, as it is difficult to bend around such a short DNA segment. The system we propose is shown in Figure 8. By varying the length of the segment containing the promoter and the length of the segment containing the terminator, we may be able to vary the ratio of determined states. ConclusionWe have evidence that short nucleotides repeats can be placed in the promoter or behind a start codon and function as randomized genetic switches through slipped-strand mutation. We have also shown that FimE can flip a segment of non-native DNA, and have evidence that the probability that FimE flips the segment depends on the length between the two binding sites. In the future, selected SSMs can be integrated into a FimE expression construct; thus, FimE expression can be randomly switched on in cells. FimE can then interact with the random state generator, depicted in Figure 8. Linked together, the probabilistic switch and the random state generator allow for the asynchronous differentiation of cells into mutually exclusive and fatal states. References |
 "
"