Team:Harvard/Dailybook/Week8/Chemical and Light
From 2008.igem.org
MIT lacZ parts RE digests 08/11
P108+45 colony PCR, P1+mtrB test digest 8/11
Nothing looks hopeful, so we'll wait until sequences come back.
Sequence results
All of the P1+mtrB not BBs aren't correct. 82 has not hits in BLAST, which 87 and 95 BLAST to pHELLSGATE- apparently some cloning vector.
Ligations/transformations 08/11
We ligated P109+63 to get RBS+full LacZ+terminator.
| Plasmid | Strain | Number of colonies |
| P63 vector control | DH5-alpha | 0 |
| P63+P109 (1:2 molar ratio) | DH5-alpha | 17 |
| P63+P109 (1:2 molar ratio) | TOP10 | 112 |
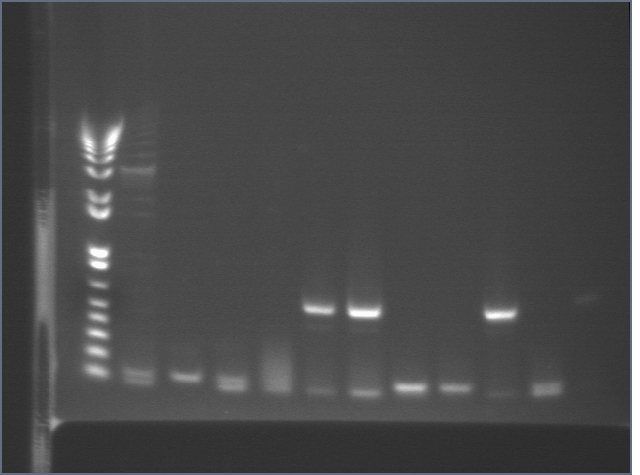
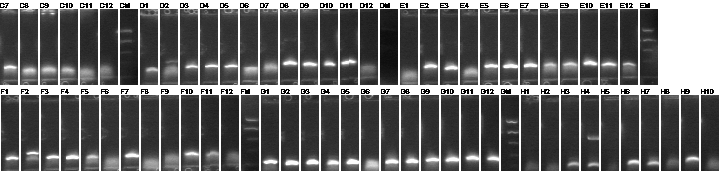
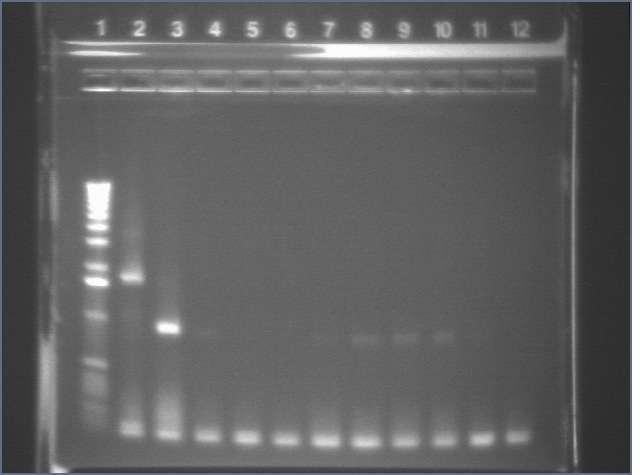
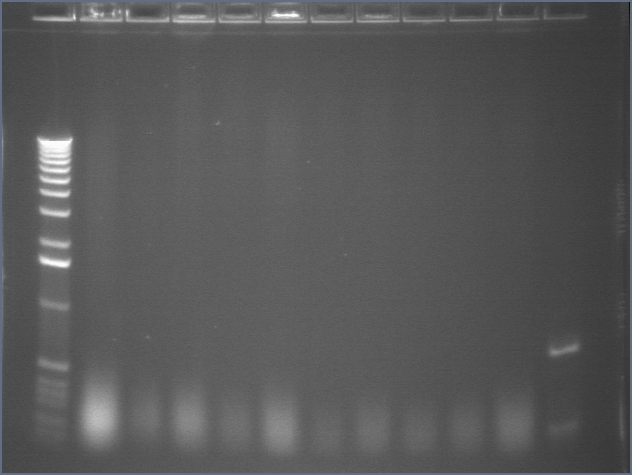
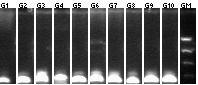
Colony PCR
We PCRed five colonies from each plate to check if they were the right size. We also used P109 as a positive control.
Lane 1: 1 kb plus ladder
Lane 2: P109 positive control (3093)
Lane 3: P109+63A (3188)
Lane 4: P109+63B (3188)
Lane 5: P109+63C (3188)
Lane 6: P109+63D (3188)
Lane 7: P109+63E (3188)
Lane 8: P109+63F (3188)
Lane 9: P109+63G (3188)
Lane 10: P109+63H (3188)
Lane 11: P109+63I (3188)
Lane 12: P109+63J (3188)
CDF
Alternative method
Using XbaI and NheI sites around the origin in pCDF, we can cut it out and ligate it into an XbaI digested P48. Depending on the orientation of the insert, the NheI-XbaI mixed site can be either just ahead of the Cm resistance (what we want) or after the E site in the BBMCS. This would destroy the prefix, so we'll use digest multiple colonies with XS for the correctly inserted origin-resistance part to put into the base pSB3K3.
pCDF, P48 digest results 8/12
The pCDF Xbal-NheI double digest (overnight, 25μL, 0.75μL of each enzyme) appeared fine, and the 800bp insert was gel purified.
P48 Xbal mysteriously yielded 2 bands, so is going to be repeated with another tube of P48.
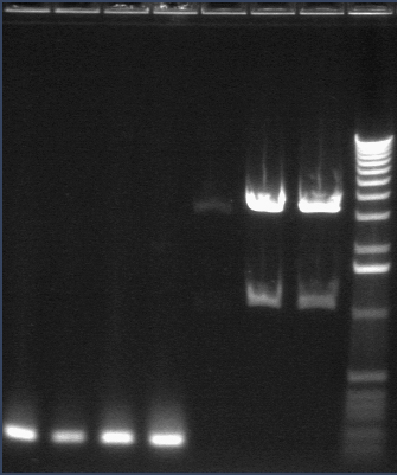
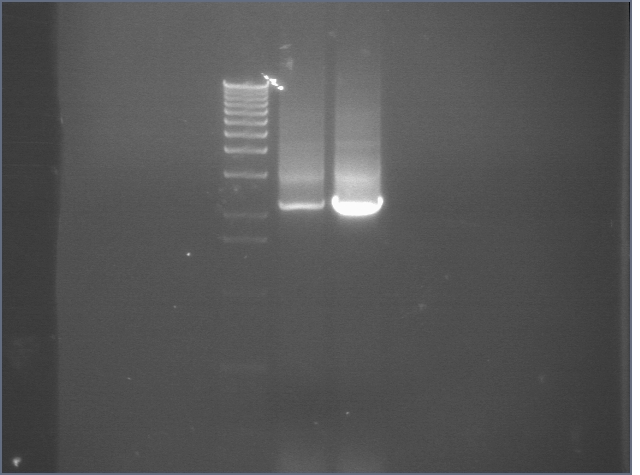
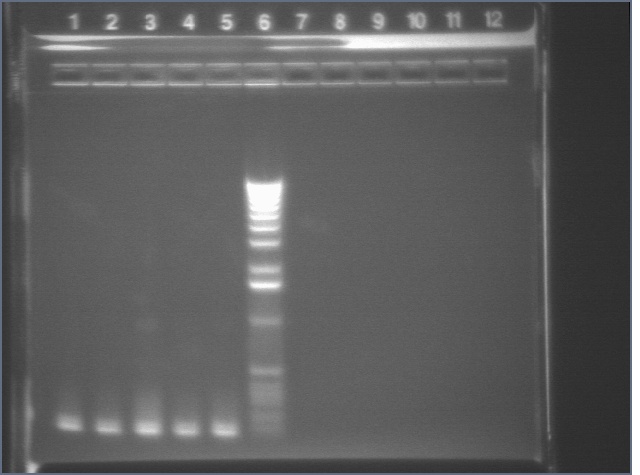
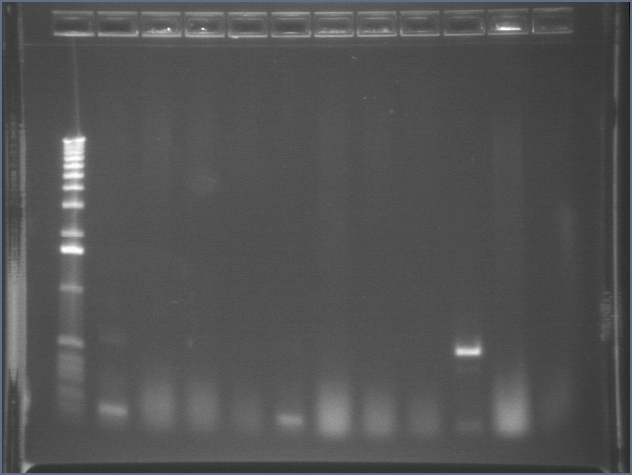
ompR (P115) digests 8/12
Since ompR #73 and #64 aligned to ompR, we're cutting it out with XP to put the whole promoter-RBS-ompR-term part into a vector with either a CDF or SC101 origin. However, these digest products were inexplicable. One had a bright 4kb band and a faint 3kb band. The other had a bright 3kb band and a very faint kb band. Although the latter is similar to what is expected, the insert was extremely faint. Digest was repeated.
Lane 1: 1 kb ladder
Lane 2: P115 uncut (~3750)
Lane 3: P115A cut XP (~1000)
Lane 4: P115B cut XP (~1000)
Lane 5: P48 uncut (3477)
Lane 6: P48 cut XbaI (3477)
PCR
8/12
Since none of the alternative strategy mtrB or P105 is working, we tried to PCR out the parts again with Phusion, as it is a more processive polymerase and our products are all >2kb.
Phusion protocol says to use Tms ≥ 60°C, so we tried both a high and low temp set for each Rx. The low temp was optimal temp from using Platinum Taq; the high temp was the highest that indicated some form of a band.
The high temperature set used as template previous iterations of the same PCR. The low temperature set used either miniprep DNA or a colony.
Rx mix: 10μL 5x HF buffer
1μL 10mM dNTPs
1.25μL each primer
0.5μL template
0.5μL Phusion polymerase
35.5 H2O
P1/3
Rx conditions: 30s @ 98°C → 5x[10s @ 98°C → 30s @ (45 or 54)°C → 1min @ 72°C] → 10x[10s @ 98°C → 30s @ (45 or 54)°C → 1min5s @ 72°C] → 7min @ 72°C → ∞ @ 4°C
45°C worked- band purified, while 54°C did not (giant smear). AK digested and column purified.
mtrB not BB
Rx conditions: 2:30s @ 98°C → 5x[10s @ 98°C → 30s @ (46 or 52)°C → 35s @ 72°C] → 10x[10s @ 98°C → 30s @ (46 or 52)°C → 40s @ 72°C] → 7min @ 72°C → ∞ @ 4°C
46°C worked- band purified, while 52°C did not (giant smear). AK digested and column purified.
mtrB not BB ligations/transformations 08/14
| Plasmid | Strain | Number of colonies |
| P1'BB vector control | DH5-alpha | 4 |
| P1+mtrB not BB | TOP10 | 24 |
| P1+mtrB not BB | DH5-alpha | 4 |
| P3 not BB vector control | DH5-alpha | 11 |
| P3+mtrB not BB | TOP10 | 104 |
| P3+mtrB notBB | DH5-alpha | 24 |
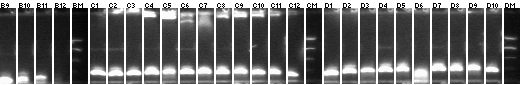
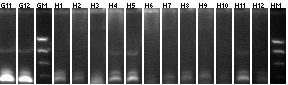
8/15 P1/3+mtrB colony PCR
PCR conditions
- 98 °C 2'30"
- Repeat 35 times
- 98 °C 10"
- 55 °C 10"
- 72 °C 45"
- 72 °C 7'
- 4 °C for ever
1-30
Lane 1: P1 vector control ligation
Lane 2: P3 vector control ligation
Lanes 3-30: P1+mtrB not BB (2.4kb)
- 10, 22 picked for mL cultures.
31-96
Lanes 31-96: P1+mtrB not BB (2.4kb)
- 44, 90 picked for 5mL cultures.
P105
P105C used for low temperature template.
Rx conditions: 2:30s @ 98°C → 5x[10s @ 98°C → 30s @ (57.8 or 60)°C → 35s @ 72°C] → 10x[10s @ 98°C → 30s @ (57.8 or 60)°C → 40s @ 72°C] → 7min @ 72°C → ∞ @ 4°C
57.8°C yielded a close double band a little bigger than 2kb (which is where the expected product is), while 60°C did not work (giant smear). We're not sure what the doublet is, so we are sending P105 for sequencing.
Redo 8/16
Template used is P105D. The expected band size is 2.2 kb. Bands excised, gel purified, AK digested, and column purified.
RE digests: Tet QPI, HO-pcyA
The P116 and P117 digests appear to be totally wrong- there should only be 1 band. The pattern resembles an incomplete digest with BB enzymes on opposite sides of the part, but we used SP.
P118 band pattern matches ~ with an incomplete digest. The insert was cut and purified.
Making Thermoinducible cI System
Ligation of P104 and P63+P18 with cI RBS from TOPO
Digestion of cI RBS from TOPO and P18+P63 8/8
P18+ P63 not confirmed by sequencing yet.
| ' | cI857 RBS TOPO ES 1 | cI857 RBS TOPO ES 2 | cI857 RBS TOPO ES 3 | cI857 RBS TOPO ES 5 | P18+ P63 ES 1 | P18+ P63 ES 2 | P18+ P63 ES 3 | P18+ P63 ES 4 | E1 P63 + RBS PP B | E1 P104 + RBS PP A | E1 P63c + RBS PP G |
| DNA | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in |
| 100X BSA | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL |
| 10X Buffer 2 | 2.5 uL | 2.5 uL | 2.5 uL | 2.5 uL | 2.5 uL | 2.5 uL | 2.5 uL | 2.5 uL | 2.5 uL of Buffer 3 | 2.5 uL of Buffer 3 | 2.5 uL of Buffer 3 |
| EcoRI | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL XbaI | 1 uL XbaI | 1 uL XbaI |
| Enzyme 2 | 1 uL SpeI | 1 uL SpeI | 1 uL SpeI | 1 uL SpeI | 1 uL XbaI | 1 uL XbaI | 1 uL XbaI | 1 uL XbaI | 1 uL PstI | 1 uL PstI | 1 uL PstI |
| Water | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in |
| Volume | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL |
Gel results 8/9
RBS TOPO 1,3, and 5, and P18+ P63 EX 1-4 were extracted and purified.
Update: At least P18+ P63 1-2 have been sequence confirmed.
Digest of P104 and P63 8/9
| ' | P63 EX | P104 EX |
| DNA | 5 uL | 15 uL |
| 100X BSA | 0.25 uL | 0.25 uL |
| 10X Buffer | 2.5 uL of Buffer 2 | 2.5 uL of Buffer 2 |
| EcoRI | 1 uL | 1 uL |
| Enzyme 2 | 1 uL XbaI | 1 uL XbaI |
| Water | 15.25 uL | 5.25 uL |
| Volume | 25 uL | 25 uL |
Gel of Digestion 8/10
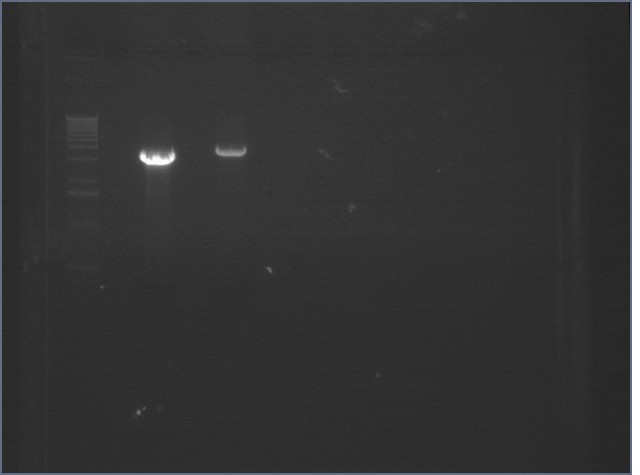
| 1% agarose gel visualized using EtBr/UV | |
|---|---|---|
| Lane | Sample | |
| 1 | 1 kB Ladder | |
| 2 | P104 EX | |
| 3 | P63 EX | |
Extracted and purified 8/10.
Ligation 8/11
Ligated P18 + P63 clones 1 and 2, and P104, with RBS from TOPO.
Plate Results 8/12
| Plate | Marker | # Colonies | Description |
| E1 P104 Dephos only | Kan | 0 colonies | |
| E1 P104 + RBS TOPO 1 | Kan | 2 0.25 mm colonies? | |
| E1 P104 + RBS TOPO 3 | Kan | 2-3 0.25mm colonies? | |
| E1 P104 + RBS TOPO 5 | Kan | 1-2 0.25mm colonies? | |
| E1 P104 + RBS TOPO 1 6:2 | Kan | 1-2 1mm colonies | |
| E1 P63 + P18 Dephos only | Amp | 0 colonies | |
| E1 P63 + P18 + RBS TOPO 1 | Amp | 1 1mm colony, 1 2mm colony | |
| E1 P63 + P18 + RBS TOPO 3 | Amp | 11 1mm colonies | not evenly distributed |
| E1 P63 + P18 + RBS TOPO 5 | Amp | 5-6 0.25mm colonies, 1 0.5mm colony | |
| E1 P63 + P18 + RBS TOPO 3 6:2 | Amp | 0 colonies | |
| E1 P63 + P18 2 Dephos Only | Amp | 2 0.25mm colonies | |
| E1 P63 + P18 2 + RBS TOPO 1 | Amp | 22 1mm colonies | Not evenly distributed. |
| E1 P63 + P18 2 + RBS TOPO 3 | Amp | ~23 1mm colonies | Not evenly distributed. |
| E1 P63 + P18 2 + RBS TOPO 5 | Amp | 16 1mm colonies | |
| Kan (-) ctrl | Kan | ~50 colonies | |
| Amp (-) ctrl | Amp | 0 colonies | |
| puc 19 (+) ctrl | Amp | 0 colonies |
Colony PCR of Transformations 8/12
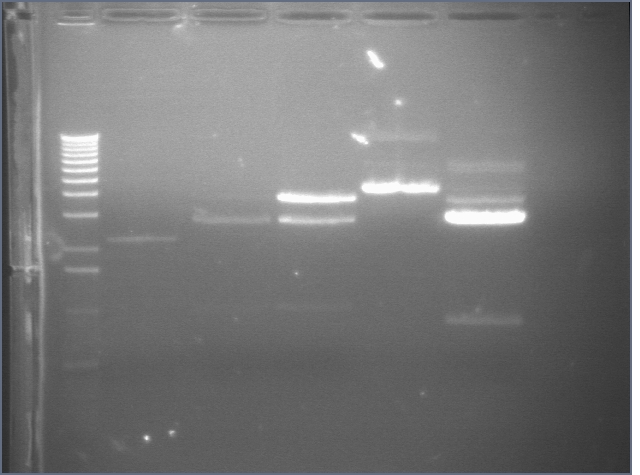
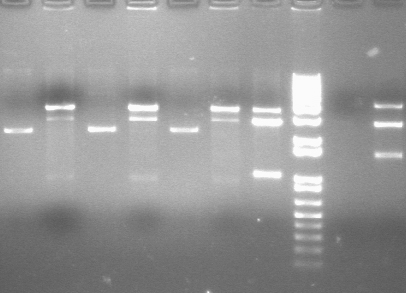
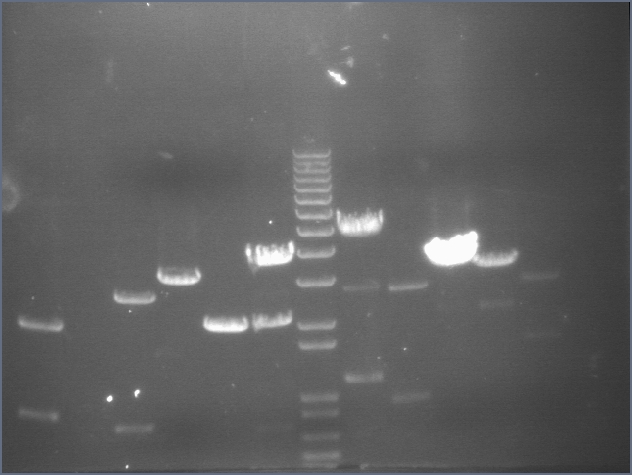
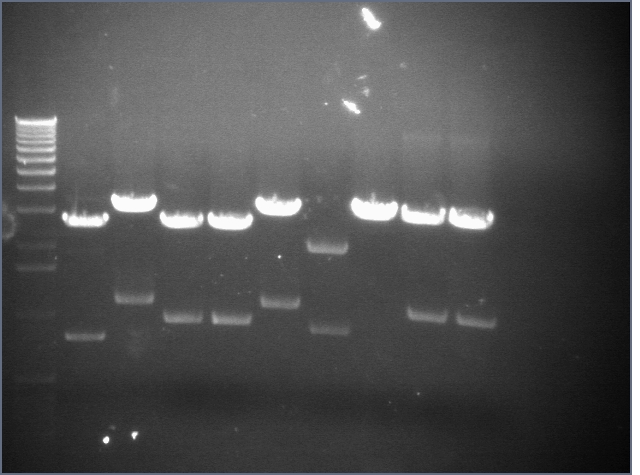
RE digests 08/14
Lane 1: P45 cut XP (876; 2079)
Lane 2: blank
Lane 3: P48 cut X (3477)
Lane 4: P63 cut EX (3284)
Lane 5: P97 cut SP (2091)
Lane 6: mtrB cut ES (~2100; 1100, 2700)
Lane 7: 1 kb plus ladder
Lane 8: mtrB cut XP (~2100; 1100, 2700)
Lane 9: P115 cut ES (~960; 2750)
Lane 10: P116 cut SP (3687)
Lane 11: P117 cut SP (3687)
Lane 12: P118 cut XP (1555; 2750)
We used the new Fermantas enzymes and digested for 6 minutes.
mtrB Ligations/transformations 08/15
Volumetric ratios not 7:1 are 2:1 molar ratios. Vector controls used plain EB to fill volume to 8μL.
| Ligation | Insert:Vector/Amount of vector (μL) | Strain | Number of colonies | Selection |
| P97 SP vector control | 0.66 | E1 | ~35 small colonies | Carb |
| mtrB(BB PCR) XP + P97 SP | 7.34:0.66 | E2 | 78 | Carb |
| mtrB(BB PCR) XP +P97 SP | 7.34:0.66 | E1 | ~20 small colonies | Carb |
| P97 SP vector control | 1 | E1 | ~25 small colonies | Carb |
| mtrB(BB PCR) XP +P97 SP | 7:1 | E1 | 8 and small colonies | Carb |
| P63 EX vector control | 3.26 | E1 | 5 | Kan |
| mtrB(TOPO) ES +63 EX | 4.74:3.26 | E2 | 36 | Kan |
| mtrB(TOPO) ES +63 EX | 4.74:3.26 | E1 | 0 | Kan |
| P63 EX vector control | 1 | E1 | 0 | Kan |
| mtrB(TOPO) ES +63 EX | 7:1 | E1 | 1 | Kan |
| mtrB(BB PCR) ES+P63 EX | 7:1 | E1 | 0 | Kan |
| P63 EX vector control | 2.37 | E1 | 0 | Kan |
| mtrB(BB PCR) ES+P63 EX | 5.63:2.37 | E1 | 1 | Kan |
| mtrB(BB PCR) ES+P63 EX | 5.63:2.37 | E2 | 128 | Kan |
Colony PCR 8/16
If mtrB is incorporated, the fragment should be ~2.2kb.
1-25 mtrB in TOPO ES + 63EX
1 picked for 5mL culture
26-60 mtrB BB PCR ES + 63EX
None look hopeful.
61-96 mtrB BB PCR XP + 97SP
62, 63, 64, 67, 85 picked for 5mL culture.
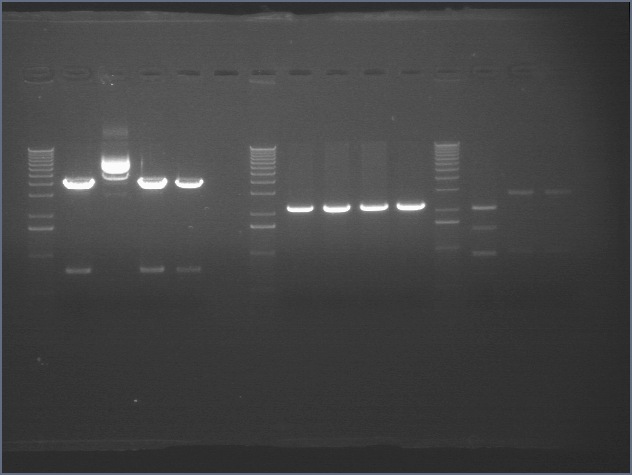
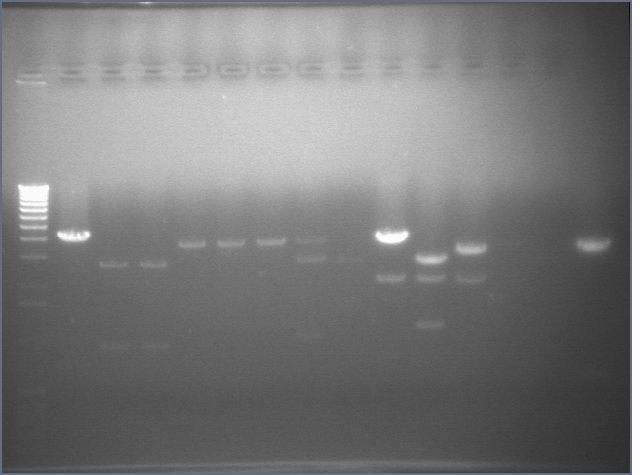
RE digests 08/15
We digested with the Fermantas enzymes for 25 minutes
Lane 1: 1 kb ladder
Lane 2: P108 cut SP (4155)
Lane 3: P115 #64 cut XP (~960; 2750)
Lane 4: P115 #73 cut XP (~960; 2750)
Lane 5: P116 #2 cut SP (3687)
Lane 6: P116 #3 cut SP (3687)
Lane 7: P116 #5 cut SP (3687)
Lane 8: P117 cut SP (3687)
Lane 9: P118 cut XP (1555; 2750)
Lane 10: mtrB TOPO cut ES (~2100; 3800)
Lane 11: mtrB TOPO cut XP (~2100; 1100, 2700)
Lane 12: P109 cut ES (3093; 2079)
Lane 13: P110 cut XP (534, 3419)
Lane 14: P111 cut XP (534, 3221)
Lane 15: P112 cut XP (534; 3189)
Ligations 8/16
Transformation results from 8/17:
(all ratios are 2:1 molar)
| Ligation | Insert | Vector | Selection | # E2 colonies | # E1 colonies |
| P97SP+mtrB topoXP | 7.26 | 0.74 | Carb | 12 | small |
| P63EX+mtrB topoES | 5.38 | 2.62 | Kan | 10 | 0 |
| P63EX+P109ES | 6.51 | 1.49 | Kan | n/a | 0 |
| P116SP+P45XP | 4.63 | 3.37 | Kan | 28 | 0 |
| P117SP+P45XP | 3.57 | 4.43 | Kan | 25 | 3 |
| P108SP+P45XP | 3.43 | 4.57 | Kan | 20 | 0 |
| P116SP+old P112XP | 2.09 | 5.91 | Kan | 70 | 2 |
| P116SP+P112XP | 3.67 | 4.33 | Kan | n/a | 0 |
| P117SP+old P112XP | 1.37 | 6.63 | Kan | n/a | 5 |
| P117SP+P112XP | 2.65 | 5.35 | Kan | n/a | 0 |
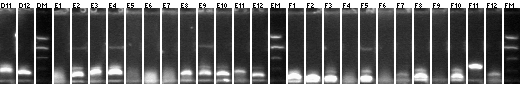
Colony PCR 08/17
Phusion polymerase used.
Master plate 1
- 1-17 P112+116
- 18-22 P112+117
- 23-42 P108+45
Master plate 2
- 1-26 P117+45
- 31-56 P116+45
Master plate 3
- 1-10 mtrB(TOPO)+P63
- 11-24 mtrB(TOPO)+P97
Strip tube PCRs (P112+116, P112+117)
Gel 1
Lane 1: 1 kb ladder
Lanes 2-12: P112+116 (Master plate 1, 1-11; 1471 kb)
9, 11 picked for 5mL cultures.
Gel 2
Lane 1: 1 kb ladder
Lanes 2-7: P112+116 (Master plate 1, 12-17; 1471 kb)
Lanes 8-12: 112+117 (Master plate 1, 18-22; 1471 kb)
21, 22 picked for 5mL cultures.
96-well plate PCRs (all the rest)
Lanes 1-20: P108+45 (Master plate 1 23-42)
30, 41, 42 picked for 5mL cultures.
Lanes 21-46: P117+45 (Master plate 2 1-26)
Lanes 47-72: P116+45 (Master plate 2 31-56)
Lanes 73-82: mtrB(TOPO)+63 (Master plate 3 1-10)
6 (lane 68) picked for 5mL culture.
Lanes 83-96: mtrB(TOPO)+97 (Master plate 3 11-24)
RE digests 08/17
We digested with Fermantas enzymes for 20 minutes.
Lane 1: 1 kb ladder
Lane 2: P48 MIT cut X (3477)
Lane 3: mtrB(BB)+P97 #63 cut XP (2100)
Lane 4: mtrB(BB)+P97 #62 cut XP (2100)
Lane 5: mtrB(BB)+P97 #64 cut XP (2100)
Lane 6: mtrB(BB)+P97 #67 cut XP (2100)
Lane 7: mtrB(BB)+P97 #85 cut XP (2100)
Lane 8: mtrB(TOPO)+P63 #1 cut XP (2200)
Lane 9: P101 cut ES (3221)
Lane 10: P101 cut XP (3221)
 "
"