Team:Heidelberg/Notebook/Killing II/8thweek
From 2008.igem.org
Revision as of 08:43, 27 October 2008 by Andreaskuehne (Talk | contribs)


8th week
Contents |
Monday 09/22/2008
pSB1A3-Receiver-Colicin cloning
- Digestion of pSB1A3-Receiver Minipreps for identification of positive clones. In addition we want to continue the colicin cloning with the cutted fragment: colonies 31, 35, 36, 39, 40, 42, 45, 66, 69, 71, 72, 73; 1h -> 37 °C
25 µl DNA 2 µl SpeI (NEB) 5 µl NEBuffer 2 (NEB) 5 µl BSA 10x (NEB) 13 µl H2O ----- 50 µl
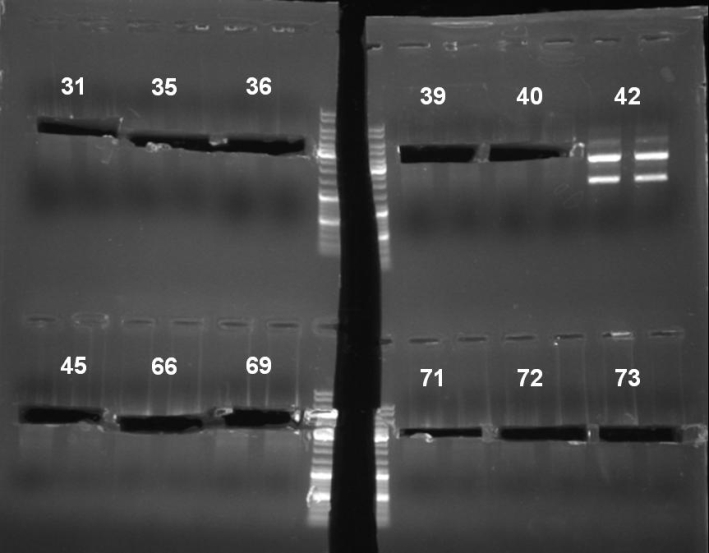
- Gel of pSB1A3-Receiver Minipreps digestion: 0.7% Agarose, 135 V, 30 min
- Gel result:
- Expected bands: pSB1A3-Receiver without GFP ~3200 bp
- There were bands at slightly different heights. We were not able to determine which of these bands have the right size. So we extracted 31, 35, 36, 39, 40, 45, 66, 69, 71, 72 and 73. To identify the right fragments we started a double digest with XbaI and SpeI. Plasmid 42 look like it was not cutted. So we expected that the fragment is ligated reverse into it.
- Gelextraction: Qiagen Kit, eluted in 40 µl H2O
- Digestion of pSB1A3-Receiver Minipreps for identification of positive clones: colonies 31, 35, 36, 39, 40, 42, 45, 66, 69, 71, 72, 73; 55 min -> 37 °C
12 µl DNA 2 µl SpeI (NEB) 2 µl XbaI (NEB) 5 µl BSA 10x (NEB) 5 µl NEBuffer 2 (NEB) 24 µl H2O ----- 50 µl
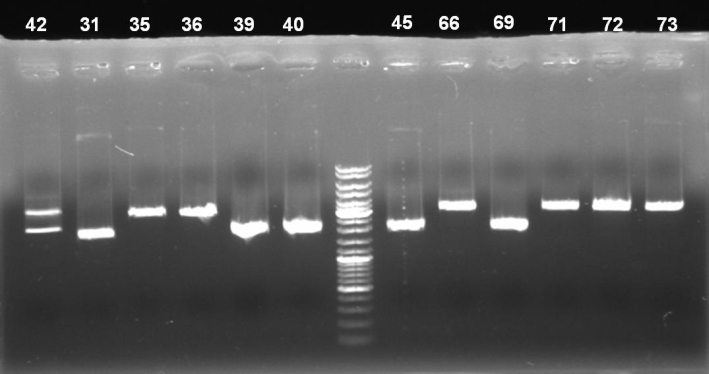
- Gel of pSB1A3-Receiver Minipreps digestion: 0.7% Agarose, 135 V, 30 min
- Gel results:
- Expected bands:
- positive clones: backbone ~2100 bp and insert ~1100 bp, partial digested ~3200
- religated clones: backbone ~2100 bp
- reverse inserted clones: no digestion
- clones with T9002 (with GFP): backbone ~2100 bp and insert (T9002) ~1950 bp
- 42: reverse inserted. To verify these assumption we send plasmid 42 to GATC for sequencing
- 31, 39, 40, 45, 69: One thick band at ~2000 bp. In the digestion with SpeI only, these plasmids had the higher bands (~3500 - 4500 bp). Because of these facts we assume that these clones contains T9002 with GFP. To verify this assumption we send plasmid 31 to GATC for sequencing.
- 35, 36, 66, 71, 72, 73: One band at ~3000. Result looks like the digestion with SpeI only. Maybe these are positive clones and we have lost the XbaI restriction site during the cloning. To verify this assumption we send plasmid 35 to GATC for sequencing.
- Expected bands:
- Digestion of Colicin E9, E9lys, E1 insert: 1h 37 °C
39 µl DNA 1 µl BamHI (NEB) 5 µl NEBuffer 3 (NEB) 5 µl BSA 10x (NEB) ----- 50 µl
- PCR Purification of Colicin E9, E9lys, E1 insert digestion (Qiagen), eluted in 30 µl
HisTag cloning of Colicins for purification
- Colony-PCR-Screen for selection of positive clones:
- ColE9: colonies 30 - 37
- ColE1: colonies 26 - 41
25.0 µl Taq Master Mic (Fermentas 2.5 µl ColE9_prot_fw_BamHI 2.5 µl ColE9_prot_rv_XmaI 20.0 µl H2O -------
program:
95 °C 3 min 95 °C 1 min | 54 °C 1 min | 30 cycles 72 °C 1 min | 72 °C 10 min 4 °C constant
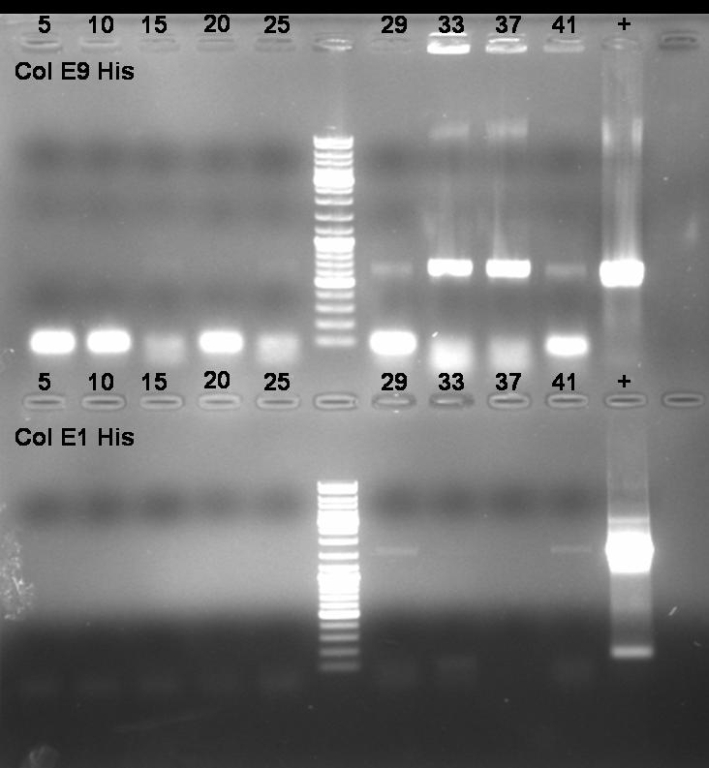
- Gel of Colony-PCR-Screen: 1% Agarose, 135V, 30 min
- Gelresults:
- ColE9His: colonies 32 and 33 have the right fragment size. Inocculation of liquid ONC.
- ColE1His: There are no colonies with the right fragment size.
Tuesday 09/23/2008
pSB1A3-Receiver-Colicin cloning
- Minipreps of pSB1A3-T9002-without GFP colonies 31, 35, 36, 39: Qiagen, Miniprepkit
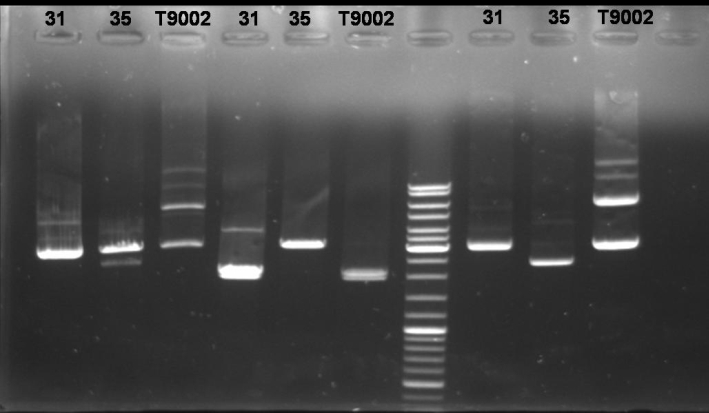
- Controldigestion of colonies 31 & 35 and BBa_T9002 with BamHI and XbaI and SpeI: 1 h 15 min -> 37 °C
- Gel of Controldigestion: 1% Agarose, 30 min, 135 V
- Gelresults:
- Colony 31: BamHI, XbaI/SpeI and undigested bands have same pattern as BBa_T9002. Maybe this is the luxpR-receiver with GFP.
- Colony 35: BamHI digestion succesful but Xba and SpeI show only one fragment like it is cutted only once.
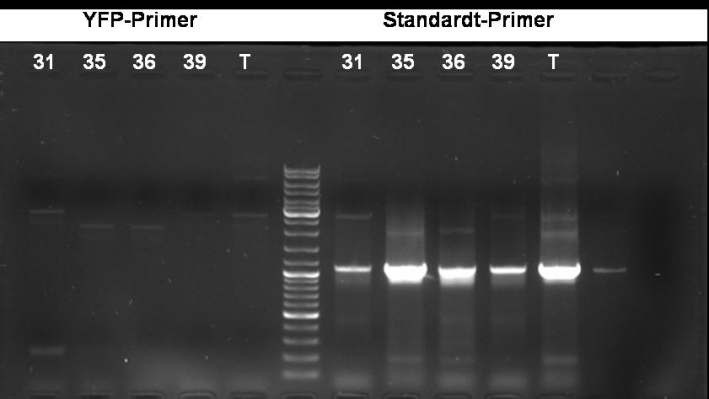
- PCR screening to check if GFP is inside the plasmid (with YFP Primers)
2.0 µl template DNA 25.0 µl Taq MasterMix 2.5 µl Primer fw 2.5 µl Primer rv 18.0 µl H2O ------- 50.0 µl
95 °C 1 min 95 °C 1 min | 56 °C 1 min | 30 cycles 72 °C 1 min | 72 °C 10 min
- Gel of PCR: No effect of YFP primers. Maybe the YFP primers binds not correctly.
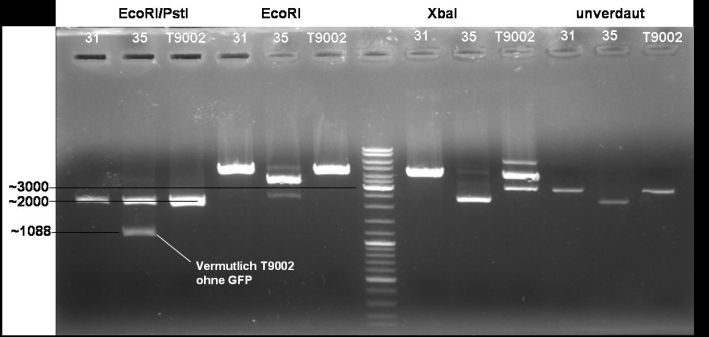
- Digestion of colonies 31, 35 and BBa_T9002 wit EcoRI/PstI, EcoRI, XbaI, undigested: 1h -> 37 °C
10/20 µl DNA 5.0 µl NEB EcoRI 5.0 µl BSA 10x 1.0 µl EcoRI 1.0 µl PstI 28/18 µl H2O -------- 50.0 µl
- Gel of digestion: 1% Agarose, 30 min, 135 V
- Gelresults: Sequencing results have shown that XbaI site is methylated at the GATC pattern. Because of that XbaI is not able to cut. To solve these problem we order new primers with the right prefix.
HisTag cloning of Colicins for purification
- PCR for selection of positive clones:
18.0 µl H2O 2.5 µl Primer fw 2.5 µl Primer rv 25.0 µl H2O ------- 50.0 µl
program 95 °C 30 sec 95 °C 1 min | 58 °C 1 min | 25 cycles 72 °C 1 min | 72 °C 10 min 4 °C constant
General
- Pascal came home from holiday: Status report
Wednesday 09/24/2008
pSB1A3-Receiver-Colicin cloning
- Sequential digestion of T9002-GFP colony 35 and ColE1.4, ColE9.4, ColE9.lys (PCR product) with SpeI and BamHI: 1h 30min -> 37 °C
25.0 µl Template DNA 5.0 µl H2O 4.0 µl NEBuffer 2/3 4.0 µl BSA10x 2.0 µl SpeI/BamHI ------- 40.0 µl
- Transformation of ColE1, ColE9 and ColE9lys (Transformation protocoll Chris)
HisTag cloning of Colicins for purification
- Sequential digestion of pQE-30 and ColE1 His (PCR product) with HindIII and BamHI: 1st step 1h 30min -> 37 °C Heatinactivation - > PCR-/Gel-Purification -> 2nd step
25.0 µl Template DNA 5.0 µl H2O 4.0 µl NEBuffer 2/3 4.0 µl BSA10x 2.0 µl HindIII/BamHI ------- 40.0 µl
- Ligation of old probes
ColE9His-pQE-30 10.0 µl pQE-30 (2.7 ng/µl) 5.0 µl ColE9His(3.3 ng/µl) 1.0 µl H2O 2.0 µl T4 DNA Lig Buffer 2.0 µl T4 DNA Ligase ------- 20.0 µl
ColE1His-pQE-30 4.5 µl pQE-30 (4.6 ng/µl) 11.5 µl ColE1His(4.4 ng/µl) 2.0 µl T4 DNA Lig Buffer 2.0 µl T4 DNA Ligase ------- 20.0 µl
neg controll ColE9His-pQE-30 10.0 µl pQE-30 (2.7 ng/µl) 6.0 µl H2O 2.0 µl T4 DNA Lig Buffer 2.0 µl T4 DNA Ligase ------- 20.0 µl
neg controll ColE1His-pQE-30 4.5 µl pQE-30 (4.6 ng/µl) 11.5 µl H2O 2.0 µl T4 DNA Lig Buffer 2.0 µl T4 DNA Ligase ------- 20.0 µl
Thursday 09/25/2008
pSB1A2-Receiver-Colicin cloning
PCR Screening colE1, colE9, colE9lys
colE1: 42 colonies picked (all) colE9 and colE9lys: 48 colonies picked
--> plated on new LB-Amp-Agar plates and PCR:
25,0 µl 2x taq MM (Fermentas) 2,5 µl Primer pSB_insert_fw 2,5 µl Primer colE1_kil_prot_rv_SpeI / colE9_plasmid_rv_SpeI / colE9_lysProt_rv_SpeI 20,0 µl H2O + picked colonies ------- 50,0 µl
95 °C 3 min _ 95 °C 1 min | 52 °C 1 min | 28x 72 °C 1 min _| 72 °C 10 min 4 °C for ever
estimated fragment lengths:
E1: 3235 bp
E9: 3143 bp
E9lys: 2167 bp
http://www.igem-heidelberg.de/fileadmin/Results/kill_2/img/080925_PCR_screening_colE1_colE9_colE9lys.jpg
HisTag cloning of Colicins for purification
- Transformation of the overnight ligation of colE1His and pQE-30 (We, 09/24/202008)
BioBrick sender part
- To create a sender part which fits BioBrick standards we want to combine BBa_F1610 with an constitutive promoter (BBa_J23107) and in addition wit a pBAD inducible promoter (BBa_I0500). Therefore we cut the promoter parts with SpeI and PstI and the sender gene cassette with XbaI and PstI. The sender gene cassette will be ligated with the cutted promoter parts.
Sender Cloning: pBAD - sender
- Digestion of BBa_I0500 with SpeI/PstI: 1h -> 37 °C
20.00 µl Miniprep DNA 1.00 µl SpeI (NEB) 0.75 µl PstI (NEB) 5.00 µl NEBuffer 2 5.00 µl BSA 10x 18.25 µl H2O -------- 50.00 µl
- Digestion of BBa_F1610 with XbaI/PstI: 1h -> 37 °C
20.00 µl Miniprep DNA 1.00 µl XbaI (NEB) 0.75 µl PstI (NEB) 5.00 µl NEBuffer 3 5.00 µl BSA 10x 18.25 µl H2O -------- 50.00 µl
- Gelextraction of cutted fragments: qiagen gelextraction kit
- Ligation of BBa_I0500 with BBa_F1610: 16 °C -> ON
15.0 µl BBa_I0500 8.0 µl BBa_F1610 1.0 µl H2O 3.0 µl T4 DNA Ligase 3.0 µl T4 DNA Ligase Buffer ------- 30.0 µl
Sender Cloning: constitutive promotor - sender
- Digestion of BBa_J23107 with SpeI/PstI: 1h -> 37 °C
20.00 µl Miniprep DNA 1.00 µl SpeI (NEB) 0.75 µl PstI (NEB) 5.00 µl NEBuffer 2 5.00 µl BSA 10x 18.25 µl H2O -------- 50.00 µl
- Digestion of BBa_F1610 with XbaI/PstI: 1h -> 37 °C
20.00 µl Miniprep DNA 1.00 µl XbaI (NEB) 0.75 µl PstI (NEB) 5.00 µl NEBuffer 3 5.00 µl BSA 10x 18.25 µl H2O -------- 50.00 µl
- Gelextraction of cutted fragments: qiagen gelextraction kit
- Ligation of BBa_I0500 with BBa_F1610: 16 °C -> ON
6.0 µl BBa_J23107 16.5 µl BBa_F1610 1.5 µl H2O 3.0 µl T4 DNA Ligase 3.0 µl T4 DNA Ligase Buffer ------- 30.0 µl
Other
- Top10 competent cells: preparation of the Top10 cultures (later continued by Kolja and Phillip)
Friday 09/26/2008
pSB1A3-Receiver-Colicin cloning
- PCR-Screen of pSB1A3-Receiver-Colicin E1/E9/E9_lys Cloning
25.0 µl Taq Master Mix (Fermentas) 2.5 µl ColE1_prot_fw_BamHI/ColE9_prot_fw_BamHI/ColE9_prot_fw_BamHI 2.5 µl ColE1_kil_prot_rv_SpeI/ColE9_prot_rv_SpeI/ColE9_lys_prot_rv_SpeI 20.0 µl H2O 5 colonies ------- 50.0 µl
program:
95 °C 3 min 95 °C 1 min | 54 °C 1 min | 30 cycles 72 °C 1 min 30 sec | 72 °C 10 min 4 °C constant
- Gel of E1 and E9 PCR-Screen: 1% Agarose, 135 V, 30 min File:080926 colE9 colE1 colony PCR screen small.jpg
- Gel-Results of E1-PCR-Screen: No colonies with excepted fragment length of ~2100 bp. But positive controll contains also not the right fragment. -> Rerun of PCR-Screen under modified conditions.
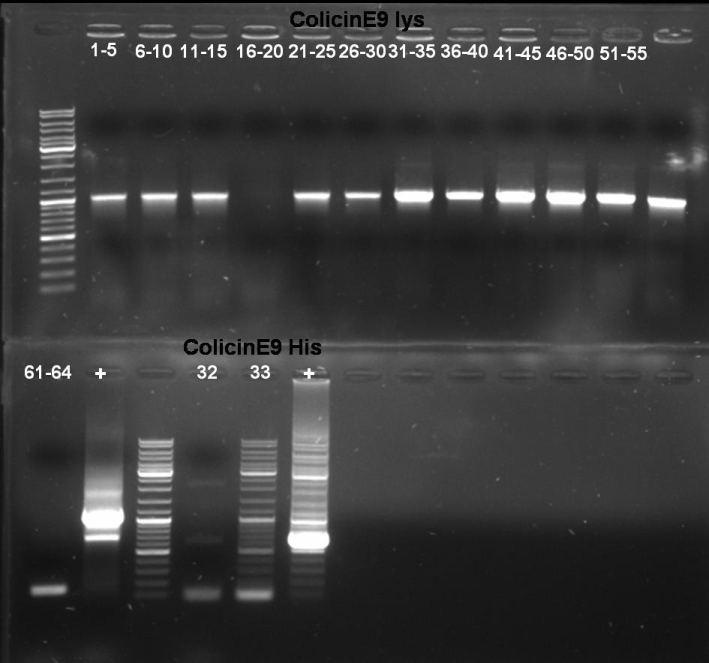
- Gel-Results of E9-PCR-Screen: Colonies 61 - 64 contains excepted fragment with ~1950 bp. -> New PCR-Screen over night of colonies 61-64. In addition: inocculation of liquid ONC of these colonies.
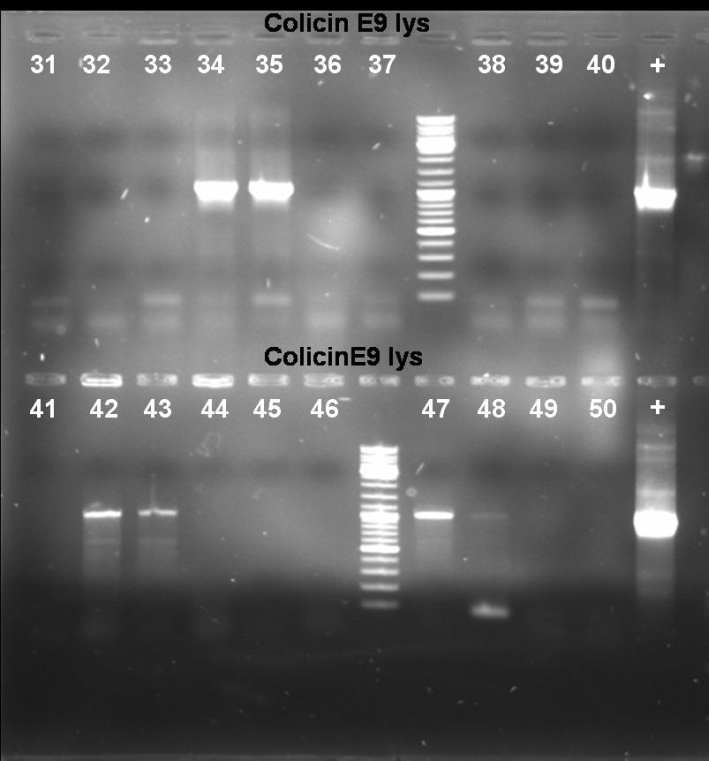
- Gel of E9lys PCR-Screen: 1% Agarose, 135 V, 30 min
- Gel-Results of E9-PCR-Screen: Colonies 1-15 and 21-60 contains excepted fragment with ~1050 bp. -> New PCR-Screen over night of colonies 31-50. In addition: inocculation of liquid ONC of these colonies.
- Overnight PCR-Screen of pSB1A3-Receiver-Colicin E1 Cloning
25.0 µl Taq Master Mix (Fermentas) 2.5 µl ColE1_prot_fw_BamHI 2.5 µl ColE1_kil_prot_rv_SpeI 20.0 µl H2O 5 colonies ------- 50.0 µl
program:
95 °C 3 min 95 °C 1 min | 65 °C 1 min | 30 cycles 72 °C 2 min | 72 °C 10 min 4 °C constant
- Overnight PCR-Screen of pSB1A3-Receiver-Colicin E9/E9lys Cloning
25.0 µl Taq Master Mix (Fermentas) 2.5 µl ColE9_prot_fw_BamHI/ColE9_prot_fw_BamHI 2.5 µl ColE9_prot_rv_SpeI/ColE9_lys_prot_rv_SpeI 20.0 µl H2O 1 colony(E9 -> 61 - 64, E9lys 31 - 50) ------- 50.0 µl
program:
95 °C 3 min 95 °C 1 min | 54 °C 1 min | 30 cycles 72 °C 2 min | 72 °C 10 min 4 °C constant
HisTag cloning of Colicins for purification
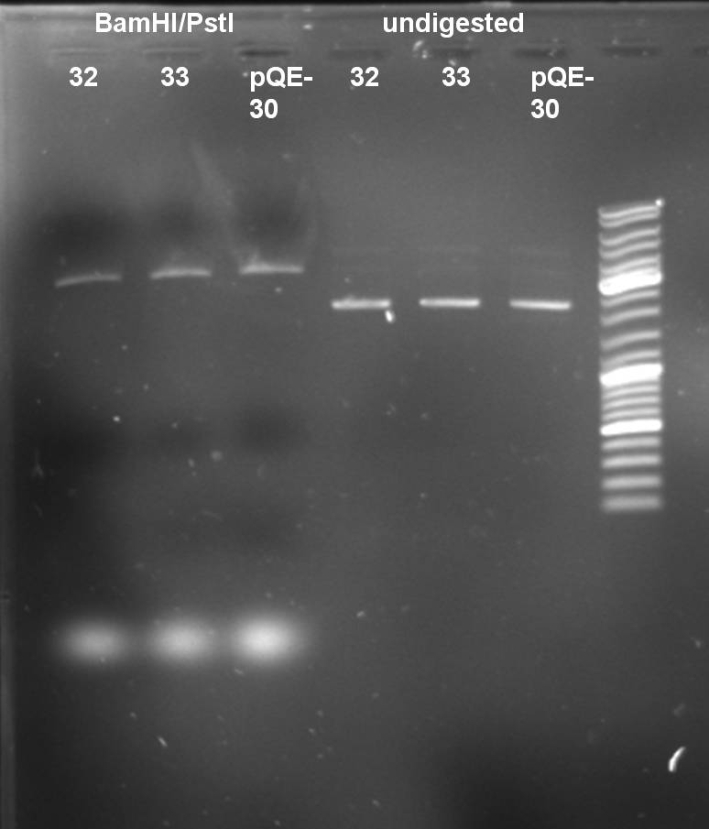
- Digestion of ColE9His 32 & 33 to verify that there is no colicin insert inside the plasmid. 1 h 37 °C
10 µl DNA (32, 33, pQE-30) 5 µl NEBuffer 3 (NEB) 5 µl BSA 10x 2 µl BamHI 2 µl PstI 26 µl H2O ----- 50 µl
- Gel of Digestion: 0.7% Agarose, 135 V, 30 min
- Results of Digestion: No 650 bp fragment visible. In addition the undigested and digested band patterns of colonies 32 & 33 look like the pattern of pQE-30. -> Cloning was not successful
Sender Cloning: pBAD - sender
- Transformation of Ligation:
Sender Cloning: constitutive promotor - sender
- Transformation of Ligation
Saturday 09/27/2008
pSB1A3-Receiver-Colicin cloning
- Sequencingresults of pSB1A3-Rec cloning:
- colony 31: contains GFP -> pSB1A3-T9002 with GFP, negative clone
- colony 35: no results available
- colony 36: contains no GFP -> positive clone
- Gel of Colony PCR Screen of pSB1A3-Receiver-ColE9lys cloning: 1% Agarose, 135 V, 30 min
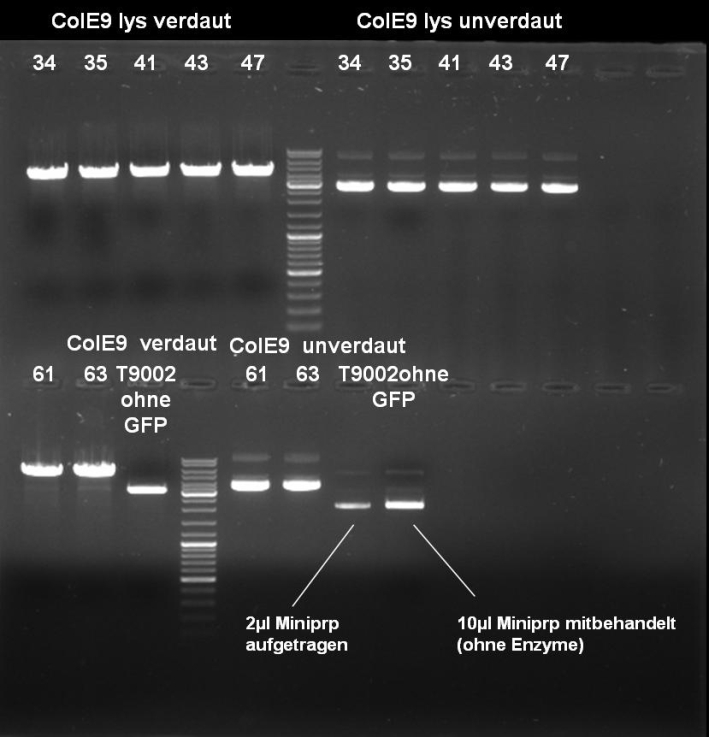
- Results of Colony PCR Screen of pSB1A3-Receiver-ColE9lys cloning: The colonies 34, 35, 42, 43 and 47 have the right fragment length (1050 bp). Miniprep of these colonies (eluted in 40 µl H2O) and glycerol stocks of colonies 34 and 35.
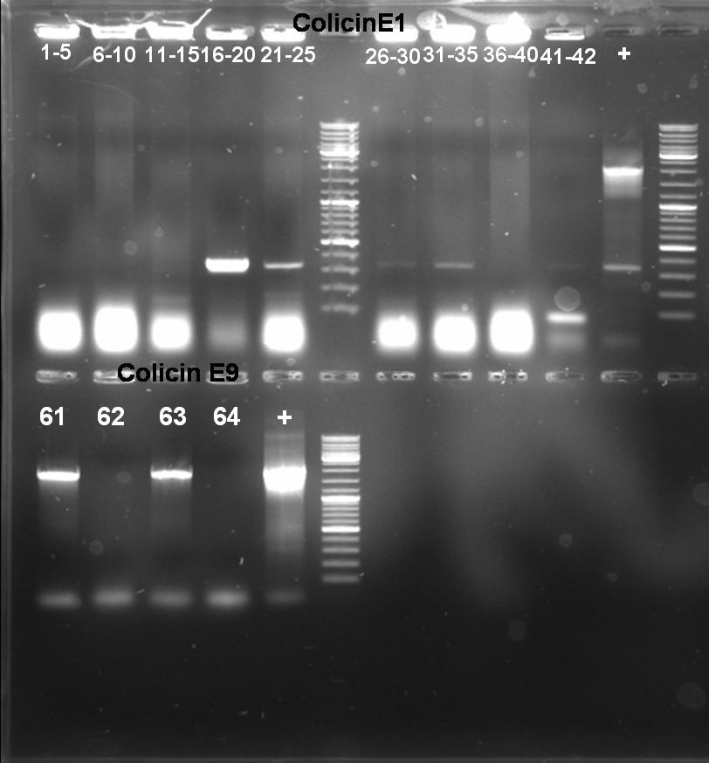
- Gel of Colony PCR Screen of pSB1A3-Receiver-ColE1 and ColE9 cloning: 1% Agarose, 135 V, 30 min
- Results of Colony PCR Screen of pSB1A3-Receiver-ColE1 cloning: No colonies with the right fragment length (~2100 bp). -> New cloning
- Results of Colony PCR Screen of pSB1A3-Receiver-ColE9 cloning: The colonies 61 and 63 have the right fragment length (~1950 bp). Miniprep of these colonies (eluted in 40 µl H2O) and glycerol stock of colony 61.
HisTag cloning of Colicins for purification
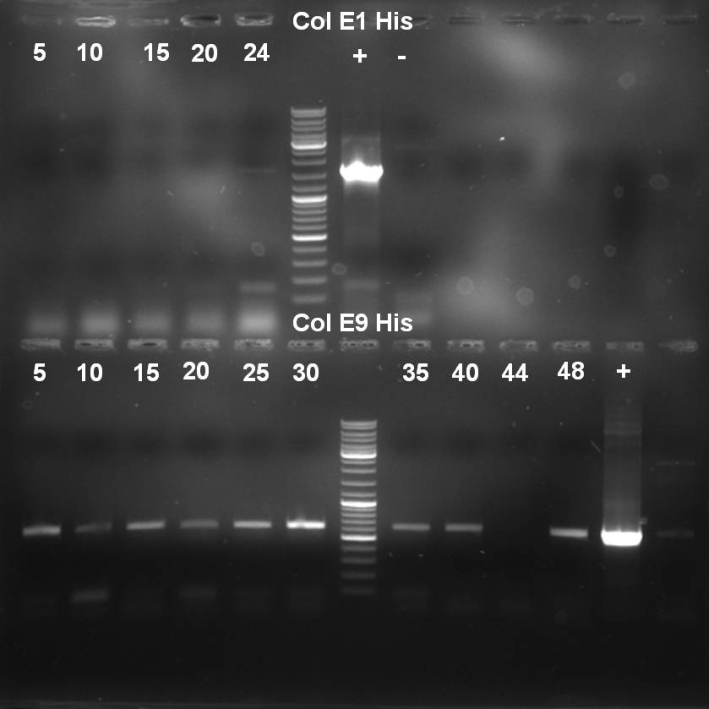
- Gel of Colony PCR Screen of Colicin E1 and E9 Histag cloning: 1% Agarose, 135 V, 30 min
- Results of HisTag-Colicin E1 Colony-PCR-Screen: No positive clones with the right fragment length (~1650 bp).
- Results of HisTag-Colicin E9 Colony-PCR-Screen: Many colonies with right fragment length (~650 bp), but the right band is also visible at the negative controll.
Sender Cloning: pBAD - sender
- Results of Transformation: one colony -> inocculation of liquid culture
Sender Cloning: constitutive promotor - sender
- Results of Transformation: two colonies -> inocculation of liquid cultures
Sunday 09/28/2008
pSB1A3-Receiver-Colicin cloning
- Digestion of Minipreps: 1h 30 min 37 °C; We did not consider the fact that our XbaI site is methylated so that only SpeI was able to cut.
10 µl DNA 1 µl XbaI (NEB) 1 µl SpeI (NEB) 5 µl BSA 10x (NEB) 5 µl NEBuffer 2(NEB) 28 µl H2O ----- 50 µl
- Gel of Digestion: 1% Agarose, 135 V, 30 min
- Ligation of pSB1A3-Receiver and Col E1: 14h at 16°C, 15 min at 65°C
7,0 µl Col E1.4 (16,5 ng/µl) 8,4 µl pSB1A3-Receiver-35 (4,1 ng/µl) 0,6 µl H2O 2,0 µl T4 DNA Ligation Buffer 2,0 µl T4 DNA Ligase ----- 20 µl
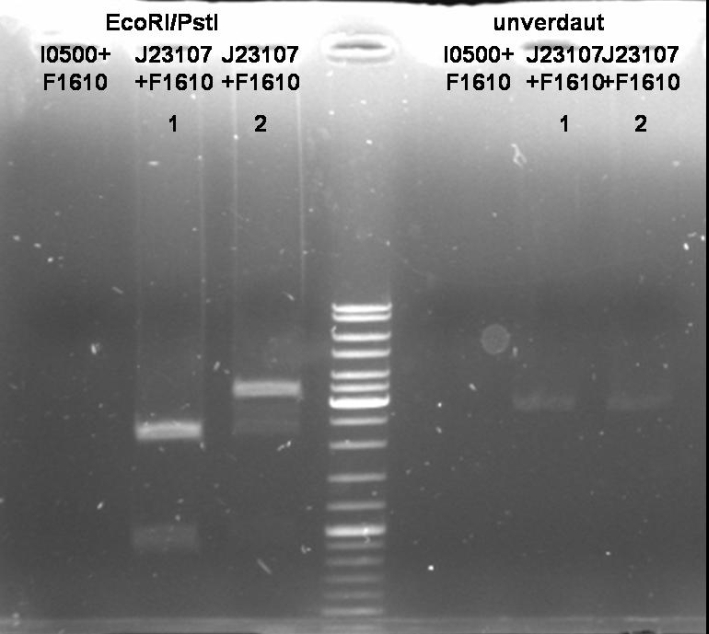
Sender Cloning: pBAD - sender
- Miniprep of ONC (Qiagen, Miniprep Kit)
- Digestion of Miniprep: 1h 30 min 37 °C
10 µl DNA 1 µl EcoRI (NEB) 1 µl PstI (NEB) 5 µl BSA 10x (NEB) 5 µl EcoRI Buffer(NEB) 28 µl H2O ----- 50 µl
- Gel of Digestion: 1% Agarose, 135 V, 30 min
Sender Cloning: constitutive promotor - sender
- Minipreps of ONC (Qiagen, Miniprep Kit)
- Digestion of Minipreps: 1h 30 min 37 °C
10 µl DNA 1 µl EcoRI (NEB) 1 µl PstI (NEB) 5 µl BSA 10x (NEB) 5 µl EcoRI Buffer(NEB) 28 µl H2O ----- 50 µl
- Gel of Digestion: 1% Agarose, 135 V, 30 min; see pBAD sender cloning
 "
"