Everything you ever wanted to know about AarI
From 2008.igem.org
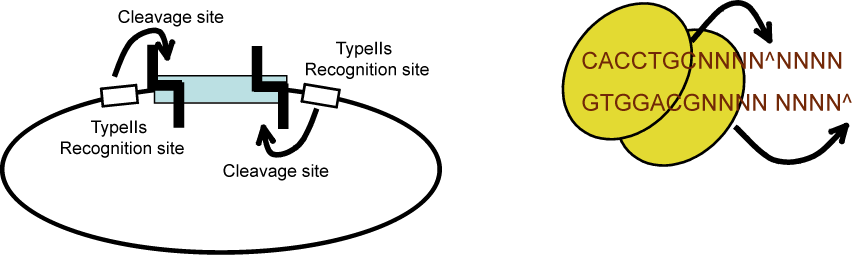
We are using a multi-part/combinatorial cloning technique that is particularly well suited to shuffling protein domains. The key to this approach is the Type IIS restriction enzyme, AarI, a rare (7-cutter) that cuts 4bp offset from its binding site. Thus, AarI can generate four base overhangs of any sequence.
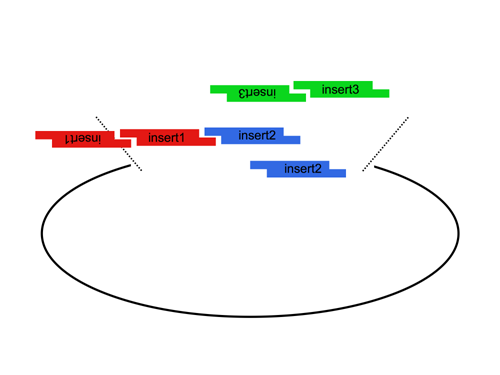
Since the user can specify the overhangs, this method can be used to "stitch-together" fragments without a scar, which is sometimes necessary to preserve protein function. More importantly, these overhangs can be non-palindromic, which solves the biggest problem faced when trying to do multipart ligations using standard restriction enzymes, the self ligation of a part, blocking it's incorporation into the construct. This problem is illustrated here:
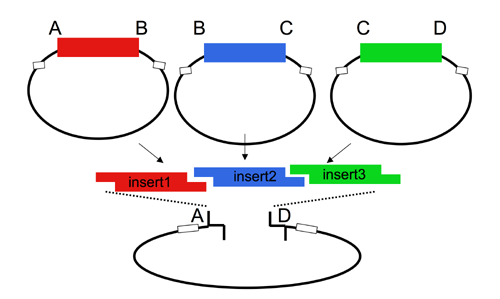
By contrast, AarI cloning allows high efficiency ligations using up to 4 parts (vector plus 3 inserts). While parts can be made with any 4 base overhang (end), we chose a standard set, termed A, B, C, and D. This allows parts to be traded between researchers. We are building a lab database of parts.
These ends yield 3 possible parts: AB, BC and CD. For two part ligations, we use AB and BD parts. Parts could be promoters, protein domains, or terminators, and are typically generated by PCR from a genomic DNA or plasmid template, then TOPO cloned and sequenced. These donor vectors, once validated, can be shuffled with other validated parts, into acceptor vectors, creating large combinatorial libraries of constructs that do not require further sequencing.
While any vector can be adapted to be an acceptor for AarI cloning, we have been working with the yeast pRS__ series of vectors ([http://www.genetics.org/cgi/reprint/122/1/19.pdf]), and we provide to the registry several types of acceptor vectors built in the pRS315 or 305 backbone. When necessary, markers can be exchanged by one-piece subclones of the completed cassettes into alternative pRS vectors, using the Kpn1/PspOMI and SacI sites.
For more information on how to clone with the UCSF AarI parts, or better yet, to design your own AarI parts, check out the following protocols.
1. Media:Design AarI primers.pdf
2. Media:How to AarI clone.pdf
== AarI Shuttle Vector ==
To facilitate exchange of parts between AarI users and the biobricks community, we are offering a Shuttle Vector. This vector accepts AarI parts in the AB or BD format (as were used in our 2008 project). These parts can then be cut out of the vector, with in-frame biobrick (SpeI/EcoRI) ends.
We are offering two versions: one that accepts AB parts (AB Shuttle) and one that accepts BD parts (BD shuttle).
AB Shuttle
GAATTC GCGGCCGC T TCTAGA ATG GGAG caaggcaggtggacaagaggagtcccgggagctggaactcccacctgcaaca CCCT T ACTAGT A GCGGCCG CTGCAG
The sequence consists of standard biobrick sites (EcoRI, NotI and XbaI at the beginnign, SpeI, NotI and PstI at the end of the acceptor sequence). There is an ATG codon (since most of our parts do not include it in their sequence), immediately followed by a sequence which includes 2 antiparallel AarI sites that produce "A" (GGAG) and "B" (CCCT) ends after digestion with AarI. Following "B" end, an additional base (T) has been introduced to keep in frame after ligating the resulting biobricks.
After insertion of the AB part aaabbbccc...zzz, the product looks like this:
...TCTAGA ATG GGAG CT aaabbbccc...zzz GGTAGTT CCCTTACTAGT...
Sequences "CT" and "GGTAGTT" that were included in our primers in all our parts.
BD Shuttle:
GAATTC GCGGCCGC T TCTAGA G CCCT caaggcaggtggacaagaggagtcccgggagctggaactcccacctgcaaca TGCG ACTAGT A GCGGCCG CTGCAG
This vector does not include start codon. The sequence with two antiparallel AarI sites is the same, the only difference is that after digestion it produces "B" (CCCT) and "D" (TGCG) ends. Again, there is an additional base (G) added before the B end to keep in frame after ligating the resulting biobricks.
After insertion of the AB part aaabbbccc...zzz, the product looks like this:
...TCTAGA G CCCT A aaabbbccc...zzz TGCG ACTAGT...
Sequence "A" after the B end was included in our primers in all our parts.
In all our BD parts we have cloned, the last triplet (zzz) is a stop codon. Therefore, biobricks derived from BD parts do not produce a fusion protein when used as front inserts. But AB parts-derived biobricks used as front inserts can be functionally ligated with biobricks derived from both AB or BD parts as back inserts. In both cases, the additional bases that have been added keep in frame. The bases were selected to generate triplets that encode aminoacids that are favored in flexible linkers.
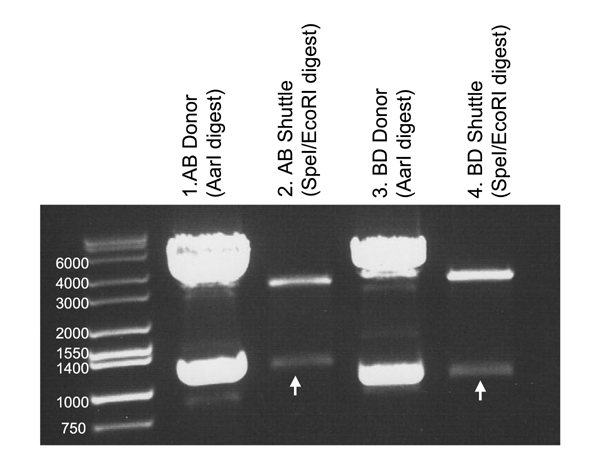
Here is an example procedure, with results, for an AB and BD fragment.
Parts were digested from donor vectors, AB and BD shuttle vectors were likewise digested (see standard AarI protocol):
AB LacI → 1107 bp part
BD RPD3cat → 1000 bp part
All fragments/vectors were gel purified, and the AB fragment was ligated into the AB shuttle, BD into the BD shuttle. Transformation efficiencies for a one-part AarI ligation are ridiculously high--it is a good idea to dilute a portion of the transformation for a lower density plate (we got lawns and had to re-streak)!
The clones obtained should contain the AarI part, now flanked by in-frame EcoRI/SpeI sites. We performed test digests to confirm:
Lane 1: The AB donor vector (LacI) was digested with AarI to release the AB fragment.
Lane 2: AB LacI was ligated into the AB Shuttle, then removed in Biobrick format by SpeI/EcoRI digest.
Lane 3: The BD donor vector (Rpd3 Catalytic domain) was digested with AarI to release the BD fragment.
Lane 4: BD Rpd3Cat was ligated into the BD Shuttle, then removed in Biobrick format by SpeI/EcoRI digest.
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Human Practices | Notebooks |
|---|
 "
"