Team:Edinburgh/Results
From 2008.igem.org
Results and current status as of 29 October 2008. We are hoping to obtain some last-minute results to present at the Jamboree.
Contents |
Overview
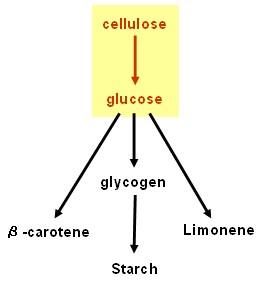
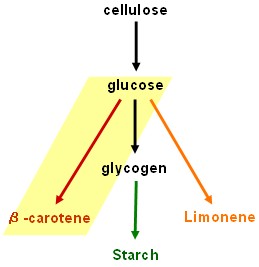
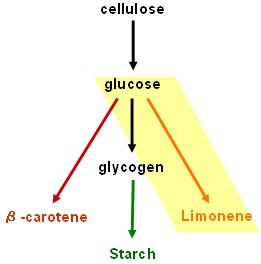
Our results are promising but our dream of using bacteria to convert cellulose into starch and β-carotene is still unrealised. Fortunately, the work will be continued by two undergraduate students after Christmas.
Cellulolysis device
- cenA (endoglucanase) and cex (exoglucanase): Biobricks made and ribosome binding site added; ready to test.
- bglX (beta glucosidase): Biobrick made and ribosome binding site added; ready to test.
- PcstA (promoter): Biobrick made and tested - successful.
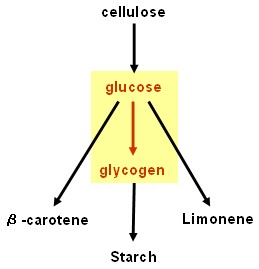
Glycogenesis device
- glgC and glgC16 (ADP-glucose pyrophosphorylase, responsible for rate limiting step in glycogenesis): Biobricks made and tested - successful.
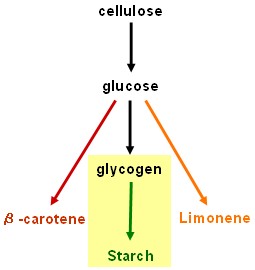
Starch synthesis device
- su1 and iso2 (isoamylases): Work in progress - pre-BioBrick constructs in pSB1A2 have been made using the BABEL protocol, but it is still necessary to remove internal EcoRI sites before fully compliant BioBricks will be ready to test. This work will be continued in an undergraduate honours project.
Lycopene generator
- dxs+crtE+crtB+crtI+crtY: Work in progress - encountered problem at final step.
Limonene synthase device
- dxs+LIMS+appY: Biobrick made - still being tested (last test failed to detect limonene synthesis).
New Protocols developed or tested
- Use of Edinbrick1 in preparation of BioBricks from PCR products
- Site-directed mutagenesis using the MABEL protocol
- BABEL: BioBrick assembly with blunt-ended ligation
- Bacillobricks: introduction of BioBricks into Bacillus subtilis
Acknowledgements
We would like to thank the following researchers for their contributions to our project:
- Raman spectroscopy for glycogen production was done by Dr Rabah Mouras.
- Mutagenesis of crtI was done by Douglas Armstrong.
- The maize isoamylase genes su1 and iso2 were generously donated by Professor Alan Myers of Iowa State University.
 "
"