Team:EPF-Lausanne/Project
From 2008.igem.org
| Home | The Team | The Project | Parts | 2-step PCR | Microfluidics | Modeling | Notebook |
|---|
Contents |
Abstract
Biological systems are unique in their ability to combine information and energy to generate complex entities. Genetically encoded networks drive many of these patterning processes. Furthermore, developmental studies have highlighted the importance of gradient formation and cell-cell communication for the generation of cellular patterns in the early stages of life. It has been shown that simple networks can form both static and dynamic patterns. Nonetheless, a system whose pattern formation is dependent on combinations of multiple signals has yet to be demonstrated. Here we address this question by designing a network, involving two different quorum-sensing based signaling mechanisms. Upon introduction in E.coli, the system can sense the relative amounts of two input molecules. Using a pre-define set of rules which was selected on its ability to generate spatial patterns, the cell can then express its final state by emitting red or green fluorescence and transmit its state to its neighbors.
General Introduction and Context
In recent years quorum sensing has been of wide-spread interest in synthetic biology. The ability for populations of cells to communicate in such a way that they either relay information or create specific patterns in culture is one which has been used in several studies (cite...). Some have relied on two different cell populations (predator-prey ref. for example) to engineer novel in vitro behavior.
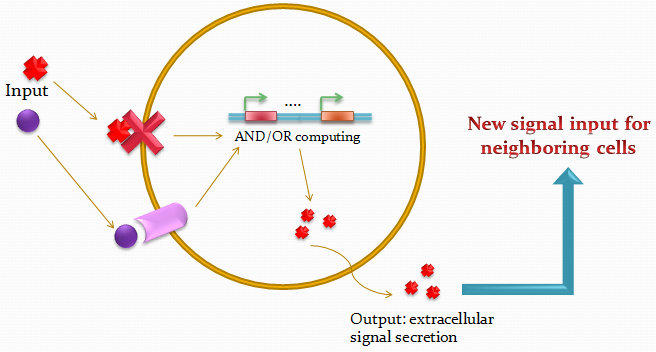
The idea of the project is to build a system which uses two different quorum sensing sets of molecules that enable the cell to integrate three different levels of signal, and emit a different response according to a predetermined rule-set, producing a given fluorescent protein and a quorum sensing molecule.
Our project uses a single population of cells, which all contain the same genetic information. Thus, the different behavior of the sub-populations will only be dictated by neighboring sub-populations.
We also take advantage of the high-level engineering competences found in our Institute (EPFL) by using microfluidic devices in order to culture the cells in an ordered and controllable manner. Furthermore this will also permit to have a control on the information flux between the cells.
In other words we want to implement an in-vivo self organising system. The cell population is self-organised in the sense that unlike other experimental setups, there is no artificial gradient
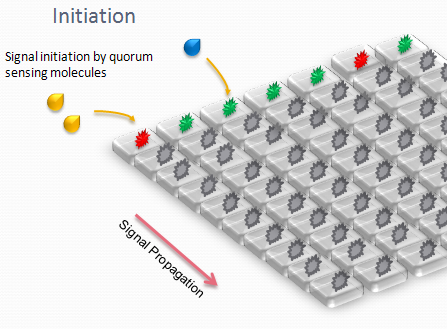
Our devices are custom-made for the project, fabricated using PDMS and multilayer soft lithography (MSL). We designed an array of miniature chambers to culture cells. The chambers are linked to one another with a network of ducts enabling us to culture sub-populations of our cells in contact with each other. Depending on the chamber connectivity and the genetic rule-set used, we expect complex patterns of GFP/RFP expression to form (see patterns below).
The aim is to initiate a signal on one side of the channel and observe the propagation along the plate.
Our microfluidic devices are also used in a side-project in order to characterize transcription factors (the MITOMI devices).
Biomedical application
Our system could be further developed to analyse blood samples, put within the cells on the first column of our chip. These cells would be enigneered to detect different molecules in blood, molecules reflecting a disorder. The system would then be able to integrate the signal across the chip. Once the signal integrated and brought to the last column, the cells placed there would be able to produce drugs to cure the disorder detected first.
Initiation of the project
Our goal was to produce a system as simple as possible giving patterns as complex as possible. We therefore decided to use only two quorum sensing systems - and two fluorescent proteins - while testing various network geometries and rule sets. We tried two different fluidic networks, determining the patterns formed by all possible rule-sets. We mean by "rule-set" the law determining the output response of the system to a given input.
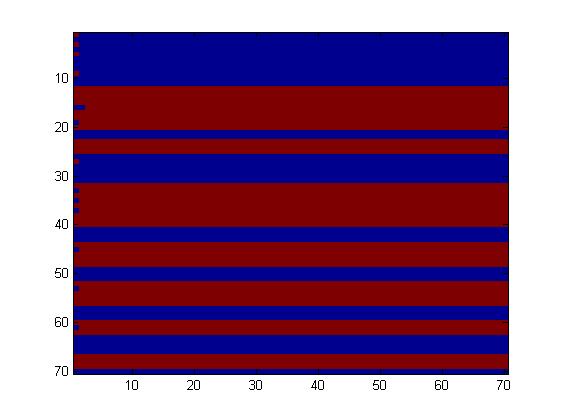
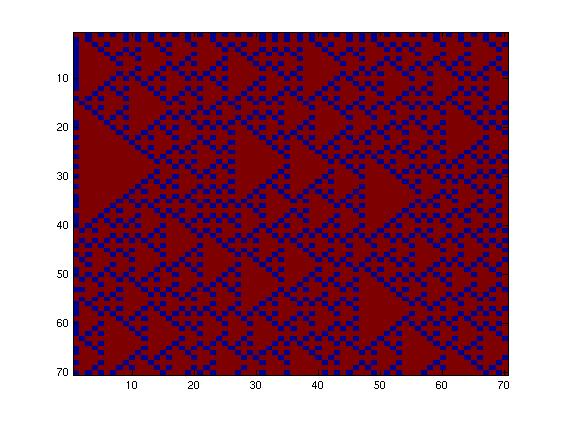
The first fluidic network, with simple connections, gave two simple patterns, like the "barcode" one.
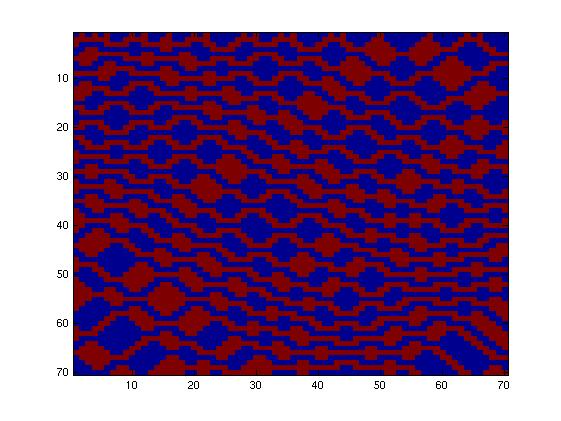
The second network, with more complex connectivity, gave more interesting patterns like the "triangles" one.
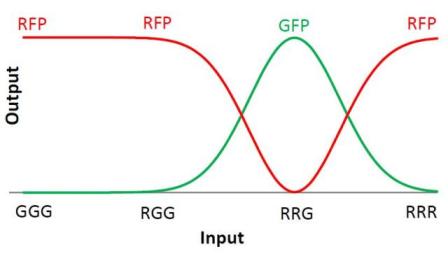
We choose to keep this pattern, and to try to implement its rule set, given in the following table. You can see the desired expression level of each fluorescent protein on the graph below.
| Input | Output |
|---|---|
| GGG | R |
| RGG | R |
| RRG | G |
| RRR | R |
The rules in this table are what we tried to implement in our system. When the cell receives a given input, it will respond by producing the correct output.
Project Details
General scheme :
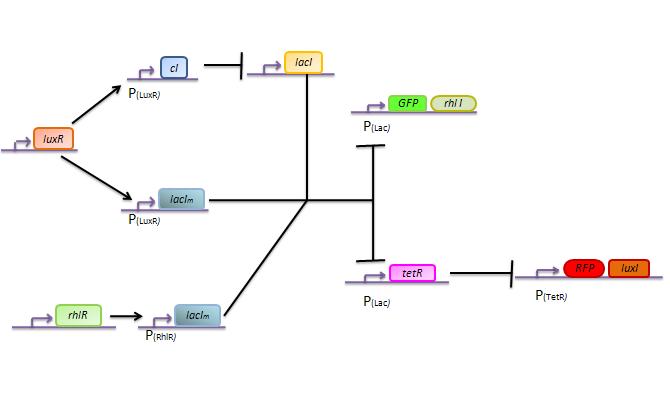
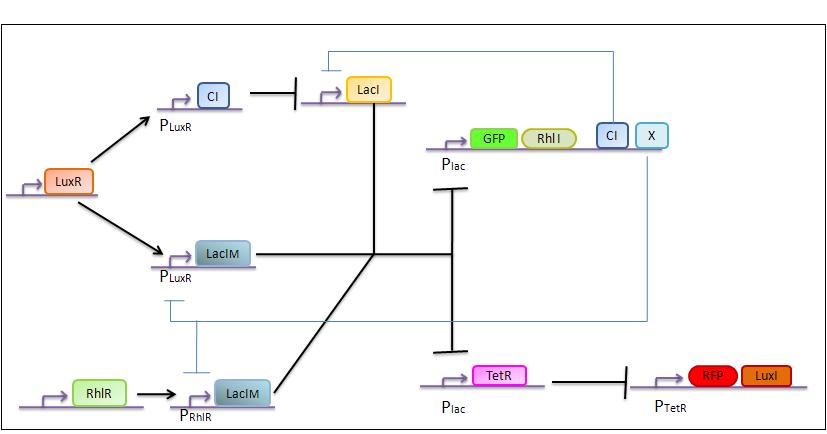
The genetic circuit which will be implemented in the cells is the following :
Pattern:
This genetic circuit must give this following pattern :
Plasmids:
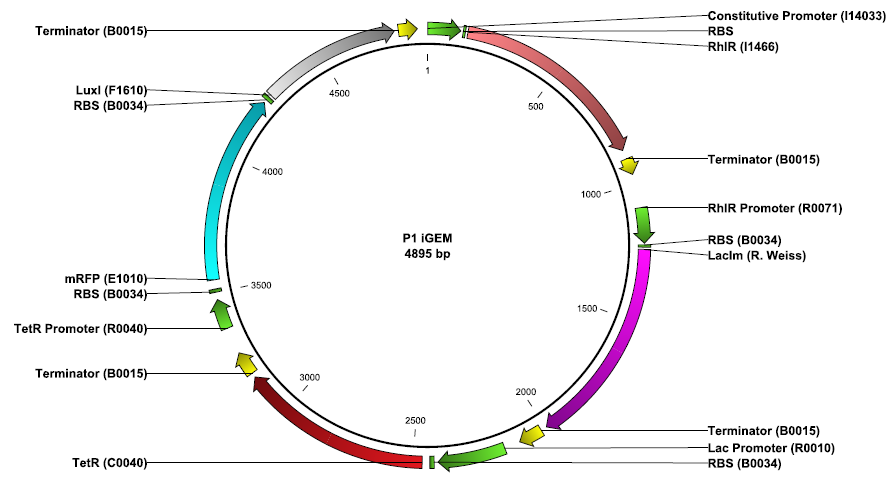
Our system is the combination between a new plasmid and a pre-existing system ("A synthetic multicellular system for programmed pattern formation", from Ron Weiss et al., Nature, vol.434, April 2005) which detect different levels of the luxI quorum sensing molecule and give a specific response according to the concentration received. As shown on the previous genetic circuit, we will add the RhlI molecule after the GFP, Rhl being our second quorum sensing molecule used. In addition to the two Ron Weiss system plasmids, we have to build an extra plasmid with the second part of the genetic circuit. We will add an extra copy of the LacIm gene so that we can test it idenpendently from the Ron Weiss system.
Here is the scheme of the plasmid with the Registry of Standard Parts references for each Biobrick we use.
Response:
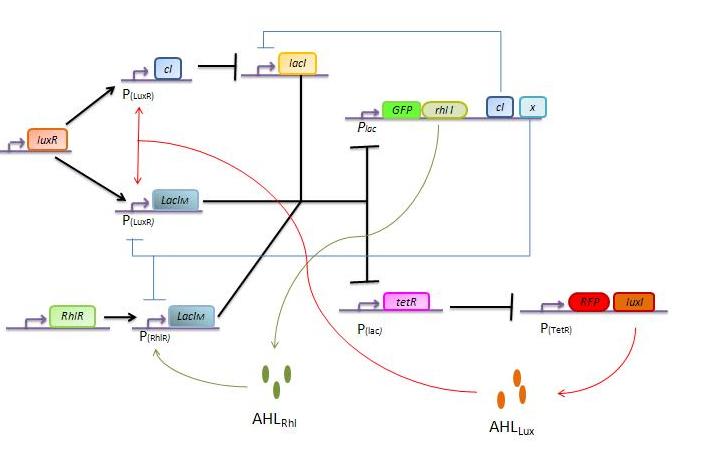
The response of our network depends on the incoming signal. Four cases can be found :
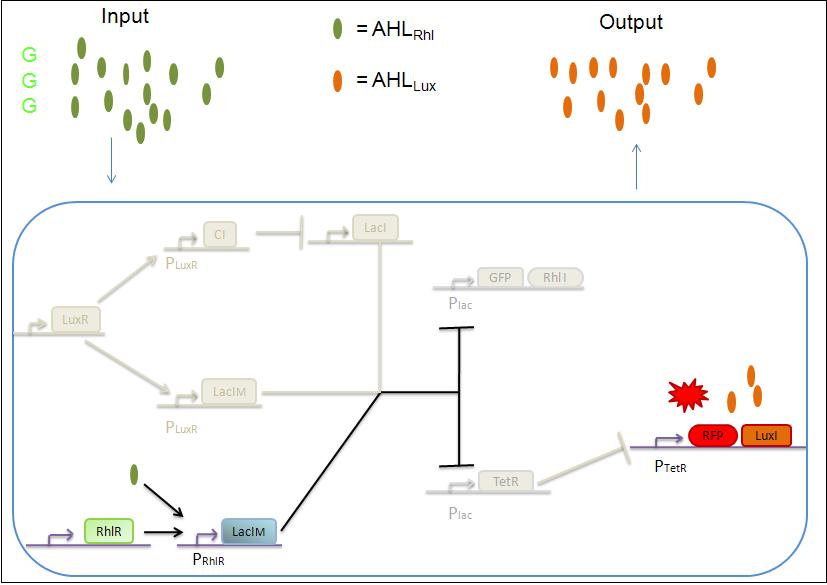
1) GGG
If there is green but no red, the AHLRhl will bind to the RhlR to allow the LacIm transcription. This LacIm will inhibit the GFP and the TetR. This latter inhibition allows the transcription of RFP and LuxI.
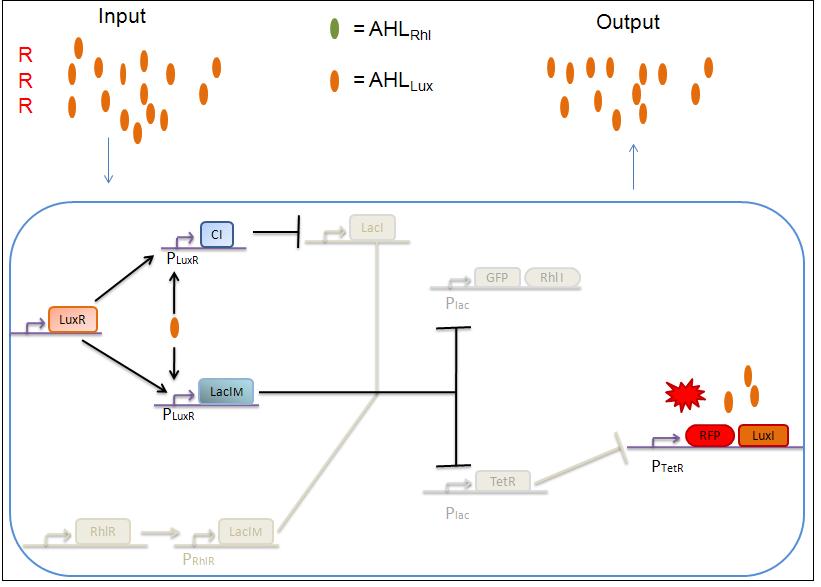
2) RRR
If there is a high level of red and no green, this means that there is a lot of AHLLux which will bind to the LuxR and allow the transcription of LacIm and Cl. Cl will then inhibit the LacI, but LacIm will inhibit on the one hand the GFP (leading to no GFP production) and on the other hand TetR. Thus, as no TetR is produced, RFP will be transcribed.
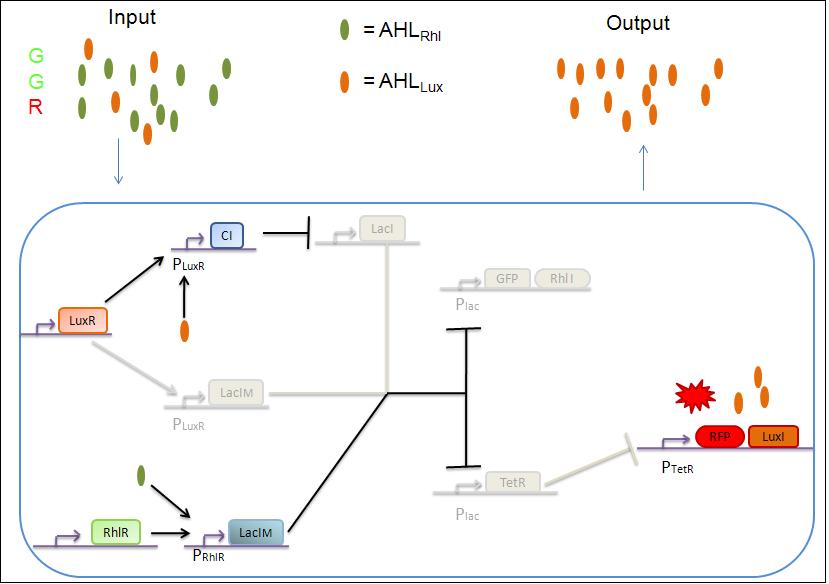
3) GGR
As both AHLRhl and AHLLux are present, we need to consider the influence of each molecule. The input of a green state means that AHLRhl will also be present, which will bind to the RhlR allowing the transcription of LacIm. This one will inhibit the GFP and also the TetR, leading to the transcription of RFP. The low input of the red state means that there is also a low level of AHLLux . This AHLLux will bind to LuxR to produce Cl and inhibit LacI. But, LacIm initiated by the RhlR promoter will remain in the cell and repress GFP and TetR, therefore leading to the expression of RFP and LuxI.
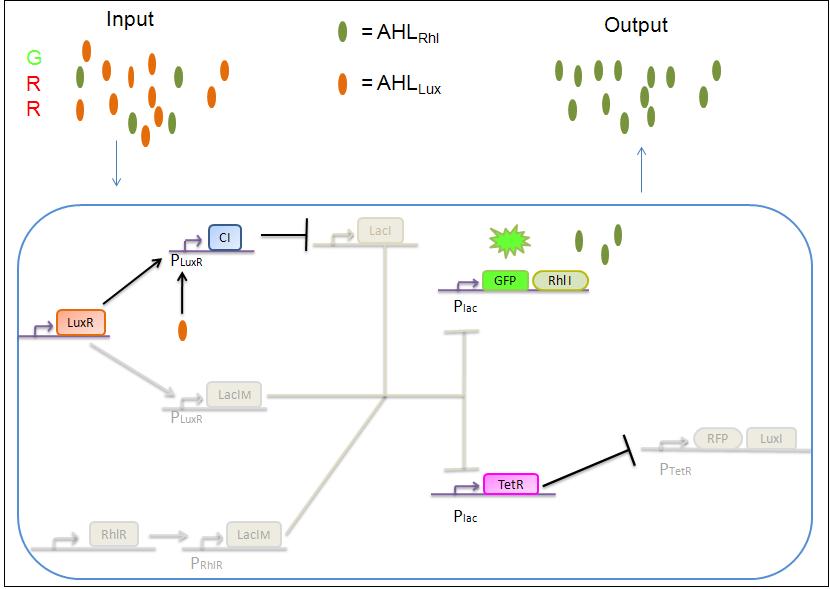
4)GRR
As both AHLRhl and AHLLux are present, we need once more to consider the influence of each molecule. Here the green input is weak and AHLRhl concentration is low. Therefore, the AHLRhl concentration will not be enough to activate LacIm. The medium input of red means that AHLLux is present in a concentration which is within the range of the band-pass. In that case, AHLLux will bind to LuxR to produce Cl but will not activate LacIm. The Cl will inhibits LacI, leading to the production of GFP and TetR. This latter will finally repress the RFP leading to a final green state with production of RhlI.
Experiments
In order to complete our project, we followed mainly three courses of action. The first focuses on the network design and the mathematical modeling; it aims to draw the principles of information processing by individual cells and the cell population, with ODEs used to model the reaction of the system. Details on this part are available in the corresponding page of the Wiki : Modeling. This part was done with the help of professor Felix Naef.
The second part of the project was of course the concrete realisation of the plasmid in the laboratory. In the facilities provided by the Laboratory of Systems Biology and Genetics (http://deplanckelab.epfl.ch/) of Prof. Bart Deplancke. Using Biobricks and material obtained from the Ron Weiss group, we followed a cloning scheme to obtain our genetic circuit in E. coli.
Most of the information on the cloning can be found on the Parts section of this Wiki, with respect to the parts submitted, on this Project page higher up (description of the plasmid), and in the Notebook section for the protocols used in culture, transformation, digestion and ligation.
The third part of the project consisted in the building and using of the microfluidic chips for the two sides of the project in the Laboratory of Biological Network Characterization (http://lbnc.epfl.ch/) of Prof. Sebastian Maerkl at EPFL. First, the MITOMI approach to used these devices in order to characterise transcription factors was implemented in the laboratory. Then, the chips for cell culture and test of the genetic circuit designed and cloned were also constructed. Information on this part of the project can be found on the corresponding page of the Wiki : Microfluidics. Protocols for the fabrication of the devices and the preparation of DNA and transcription factors can also be found on the Notebook page of this Wiki.
Results
We were able to obtain the cloned plasmid with all the components.
Several MITOMI experiments were conducted with the chips.
We really need more detail on all of this, including Keshav's fluorescence measurements. We can link to the significant steps of the cloning in the Notebook part as well.
 "
"