Team:Harvard/Dailybook/Week5/Chemical and Light
From 2008.igem.org
Testing Thermoinducible Lac System
Restreaked Plates of P92-3 in S1 7/21
Restreaked P93 (1:3), P92a, and P93b--all successfully transformed in S1--to detect YFP at 30 and 40 degrees.
Results 7/22
S1 P93 (1:3), P92a, and P93b plates grown at 30 and 40 degrees failed to demonstrate any difference in YFP expression. Both expressed very low, if any, YFP.
Thermoinducible Test 7/24
| ' | ' | ' | Time | ' | ' | ' | ' | ' | ' | ' | ' | ' | ' | ' | ' |
| 0 hrs | 2 hrs | 4 hrs | 6 hrs | ||||||||||||
| Strain | Replicate | Before Splitting | 30 | 30 + 1mM IPTG | 40 | 42 | 30 | 30 + 1mM IPTG | 40 | 42 | 30 | 30 + 1mM IPTG | 40 | 42* DID NOT SHAKE | |
| LB | N/A | Dilution | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| OD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| YFP | 36.78 | 40.72 | 40.72 | 40.72 | 40.72 | 41.32 | 41.32 | 41.32 | 41.32 | 45.4 | 45.4 | 45.4 | 45.4 | ||
| P84 | A | Dilution | N/A | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | 1 in 5 | 1 in 5 | 1 in 5.5 | 1 in 5.5 | 1 in 6 | 1 in 5.5 | 1 in 6.5 | 1 in 5.5 |
| OD | 0.269 | 0.229 | 0.218 | 0.216 | 0.214 | 0.241 | 0.224 | 0.227 | 0.219 | 0.243 | 0.234 | 0.243 | 0.224 | ||
| YFP | 291.95 | 242.89 | 226.53 | 216.85 | 211.21 | 226.95 | 222.07 | 196.83 | 196.3 | 250.22 | 238.06 | 224.98 | 215.35 | ||
| YFP/OD | 1085.315985 | 1060.655022 | 1039.12844 | 1003.935185 | 986.9626168 | 941.7012448 | 991.3839286 | 867.092511 | 896.347032 | 1029.711934 | 1017.350427 | 925.8436214 | 961.3839286 | ||
| B | Dilution | N/A | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | 1 in 5 | 1 in 5 | 1 in 5.5 | 1 in 5.5 | 1 in 6 | 1 in 5.5 | 1 in 6.5 | 1 in 5.5 | |
| OD | 0.269 | 0.221 | 0.218 | 0.203 | 0.208 | 0.231 | 0.228 | 0.221 | 0.228 | 0.233 | 0.23 | 0.228 | 0.268 | ||
| YFP | 291.95 | 225.35 | 229.19 | 208.3 | 199.34 | 215.49 | 208.55 | 195.39 | 192.62 | 233.83 | 233.99 | 215.07 | 241.39 | ||
| YFP/OD | 1085.315985 | 1019.683258 | 1051.330275 | 1026.108374 | 958.3653846 | 932.8571429 | 914.6929825 | 884.1176471 | 844.8245614 | 1003.562232 | 1017.347826 | 943.2894737 | 900.7089552 | ||
| C | Dilution | N/A | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | 1 in 5 | 1 in 5 | 1 in 5.5 | 1 in 5.5 | 1 in 6 | 1 in 5.5 | 1 in 6.5 | 1 in 5.5 | |
| OD | 0.269 | 0.239 | 0.225 | 0.22 | 0.203 | 0.24 | 0.218 | 0.225 | 0.225 | 0.231 | 0.244 | 0.236 | 0.237 | ||
| YFP | 291.95 | 243.21 | 233.06 | 207.05 | 195.54 | 221.06 | 202.99 | 193.67 | 188.6 | 233.191 | 242.86 | 219.04 | 218.02 | ||
| YFP/OD | 1085.315985 | 1017.615063 | 1035.822222 | 941.1363636 | 963.2512315 | 921.0833333 | 931.146789 | 860.7555556 | 838.2222222 | 1009.484848 | 995.3278689 | 928.1355932 | 919.9156118 | ||
| P85 | A | Dilution | N/A | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | 1 in 5 | 1 in 5 | 1 in 5.5 | 1 in 5.5 | 1 in 6 | 1 in 5.5 | 1 in 6.5 | 1 in 5.5 |
| OD | 0.221 | 0.227 | 0.218 | 0.2 | 0.196 | 0.232 | 0.226 | 0.212 | 0.217 | 0.239 | 0.234 | 0.218 | 0.221 | ||
| YFP | 245.2 | 236.07 | 228.69 | 202.98 | 190.63 | 220.43 | 208.21 | 184.71 | 187.91 | 239.59 | 236.01 | 212.75 | 209.69 | ||
| YFP/OD | 1109.502262 | 1039.955947 | 1049.036697 | 1014.9 | 972.6020408 | 950.1293103 | 921.2831858 | 871.2735849 | 865.9447005 | 1002.468619 | 1008.589744 | 975.9174312 | 948.8235294 | ||
| B | Dilution | N/A | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | 1 in 5 | 1 in 5 | 1 in 5.5 | 1 in 5.5 | 1 in 6.5 | 1 in 5.5 | 1 in 6.5 | 1 in 5.5 | |
| OD | 0.221 | 0.233 | 0.219 | 0.202 | 0.192 | 0.244 | 0.223 | 0.213 | 0.22 | 0.229 | 0.225 | 0.225 | 0.225 | ||
| YFP | 245.2 | 232.52 | 224.04 | 200.66 | 190.6 | 220 | 204.88 | 181.33 | 179.51 | 233.89 | 227.6 | 211.73 | 209.61 | ||
| YFP/OD | 1109.502262 | 997.9399142 | 1023.013699 | 993.3663366 | 992.7083333 | 901.6393443 | 918.7443946 | 851.314554 | 815.9545455 | 1021.353712 | 1011.555556 | 941.0222222 | 931.6 | ||
| C | Dilution | N/A | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | 1 in 5 | 1 in 5 | 1 in 5.5 | 1 in 5.5 | 1 in 6 | 1 in 6 | 1 in 6.5 | 1 in 5.5 | |
| OD | 0.221 | 0.23 | 0.226 | 0.2 | 0.199 | 0.238 | 0.229 | 0.216 | 0.217 | 0.238 | 0.232 | 0.234 | 0.227 | ||
| YFP | 245.2 | 233.88 | 229.97 | 199.54 | 193.24 | 215.08 | 208.23 | 182.57 | 182.58 | 234.45 | 232.47 | 217.81 | 210.75 | ||
| YFP/OD | 1109.502262 | 1016.869565 | 1017.566372 | 997.7 | 971.0552764 | 903.697479 | 909.30131 | 845.2314815 | 841.3824885 | 985.0840336 | 1002.025862 | 930.8119658 | 928.4140969 | ||
| (-) Ctrl (P41) | A | Dilution | N/A | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | 1 in 5 | 1 in 5 | 1 in 5.5 | 1 in 5.5 | 1 in 6 | 1 in 5.5 | 1 in 6.5 | 1 in 5.5 |
| OD | 0.216 | 0.232 | 0.215 | 0.197 | 0.199 | 0.237 | 0.221 | 0.217 | 0.223 | 0.234 | 0.237 | 0.226 | 0.234 | ||
| YFP | 234.27 | 227.67 | 216.53 | 186.19 | 198.19 | 214.86 | 204.3 | 190.44 | 205.31 | 236.74 | 234.19 | 211.12 | 215.13 | ||
| YFP/OD | 1084.583333 | 981.3362069 | 1007.116279 | 945.1269036 | 995.9296482 | 906.5822785 | 924.4343891 | 877.6036866 | 920.6726457 | 1011.709402 | 988.1434599 | 934.159292 | 919.3589744 | ||
| B | Dilution | N/A | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | 1 in 5 | 1 in 5 | 1 in 5.5 | 1 in 5.5 | 1 in 6 | 1 in 6 | 1 in 6.5 | 1 in 5.5 | |
| OD | 0.216 | 0.227 | 0.234 | 0.199 | 0.197 | 0.235 | 0.223 | 0.217 | 0.219 | 0.242 | 0.233 | 0.225 | 0.228 | ||
| YFP | 234.27 | 237.44 | 226.99 | 190.07 | 186.35 | 215.29 | 210.32 | 190.44 | 188.45 | 239.24 | 234.96 | 209.83 | 207.08 | ||
| YFP/OD | 1084.583333 | 1045.991189 | 970.042735 | 955.1256281 | 945.9390863 | 916.1276596 | 943.1390135 | 877.6036866 | 860.5022831 | 988.5950413 | 1008.412017 | 932.5777778 | 908.245614 | ||
| C | Dilution | N/A | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | 1 in 5 | 1 in 5 | 1 in 5.5 | 1 in 5.5 | 1 in 6 | 1 in 6 | 1 in 6.5 | 1 in 5.5 | |
| OD | 0.216 | 0.219 | 0.218 | 0.193 | 0.194 | 0.238 | 0.228 | 0.214 | 0.219 | 0.241 | 0.229 | 0.22 | 0.223 | ||
| YFP | 234.27 | 224.51 | 227.38 | 201.61 | 190.81 | 216.18 | 209.61 | 185.68 | 183.13 | 240.67 | 227.78 | 208.24 | 207.36 | ||
| YFP/OD | 1084.583333 | 1025.159817 | 1043.027523 | 1044.611399 | 983.556701 | 908.3193277 | 919.3421053 | 867.6635514 | 836.2100457 | 998.6307054 | 994.6724891 | 946.5454545 | 929.8654709 |
Data for the 1000fold Dilutions after 2 Hours
| ' | ' | ' | Time | ' | ' | ' | ' | ' | ' | ' | ' | ' |
| 2hrs | Diluted 1000 fold, grown for 5 hours. | Diluted 1000 fold, grown for 5 hours, diluted to same OD range. | ||||||||||
| Strain | Replicate | 30 | 30 + 1mM IPTG | 40 | 42 | 30 | 30 + 1mM IPTG | 40 | 42* DID NOT SHAKE FOR TWO HOURS IN THE MIDDLE. | 40 | 42* DID NOT SHAKE FOR TWO HOURS IN THE MIDDLE. | |
| LB | N/A | Dilution | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 1 in 10 | 1 in 10 |
| OD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| YFP | 40.72 | 40.72 | 40.72 | 40.72 | 46.8 | 46.8 | 46.8 | 46.8 | 46.8 | 46.8 | ||
| P84 | A | Dilution | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | N/A | N/A | N/A | N/A | 1 in 10 | 1 in 10 |
| OD | 0.229 | 0.218 | 0.216 | 0.214 | 0.018 | 0.015 | 0.12 | 0.139 | 0.015 | 0.018 | ||
| YFP | 242.89 | 226.53 | 216.85 | 211.21 | 63.52 | 59.56 | 164.97 | 169.15 | 59.29 | 61.78 | ||
| YFP/OD | 1060.655022 | 1039.12844 | 1003.935185 | 986.9626168 | 3528.888889 | 3970.666667 | 1374.75 | 1216.906475 | 3952.666667 | 3432.222222 | ||
| B | Dilution | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | N/A | N/A | N/A | N/A | 1 in 10 | 1 in 10 | |
| OD | 0.221 | 0.218 | 0.203 | 0.208 | 0.015 | 0.016 | 0.134 | 0.127 | 0.02 | 0.018 | ||
| YFP | 225.35 | 229.19 | 208.3 | 199.34 | 62.13 | 63.2 | 169.54 | 170.49 | 61.6 | 62.48 | ||
| YFP/OD | 1019.683258 | 1051.330275 | 1026.108374 | 958.3653846 | 4142 | 3950 | 1265.223881 | 1342.440945 | 3080 | 3471.111111 | ||
| C | Dilution | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | N/A | N/A | N/A | N/A | 1 in 10 | 1 in 10 | |
| OD | 0.239 | 0.225 | 0.22 | 0.203 | 0.017 | 0.016 | 0.131 | 0.127 | 0.021 | 0.016 | ||
| YFP | 243.21 | 233.06 | 207.05 | 195.54 | 60.86 | 62.2 | 164.97 | 163.9 | 59.74 | 64.82 | ||
| YFP/OD | 1017.615063 | 1035.822222 | 941.1363636 | 963.2512315 | 3580 | 3887.5 | 1259.312977 | 1290.551181 | 2844.761905 | 4051.25 | ||
| P85 | A | Dilution | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | N/A | N/A | N/A | N/A | 1 in 10 | 1 in 10 |
| OD | 0.227 | 0.218 | 0.2 | 0.196 | 0.014 | 0.017 | 0.121 | 0.122 | 0.018 | 0.016 | ||
| YFP | 236.07 | 228.69 | 202.98 | 190.63 | 59.28 | 62.21 | 155.08 | 173.29 | 63.34 | 61.37 | ||
| YFP/OD | 1039.955947 | 1049.036697 | 1014.9 | 972.6020408 | 4234.285714 | 3659.411765 | 1281.652893 | 1420.409836 | 3518.888889 | 3835.625 | ||
| B | Dilution | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | N/A | N/A | N/A | N/A | 1 in 10 | 1 in 10 | |
| OD | 0.233 | 0.219 | 0.202 | 0.192 | 0.014 | 0.012 | 0.121 | 0.114 | 0.018 | 0.016 | ||
| YFP | 232.52 | 224.04 | 200.66 | 190.6 | 60.47 | 56.56 | 149.74 | 175 | 60.99 | 57.54 | ||
| YFP/OD | 997.9399142 | 1023.013699 | 993.3663366 | 992.7083333 | 4319.285714 | 4713.333333 | 1237.520661 | 1535.087719 | 3388.333333 | 3596.25 | ||
| C | Dilution | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | N/A | N/A | N/A | N/A | 1 in 10 | 1 in 10 | |
| OD | 0.23 | 0.226 | 0.2 | 0.199 | 0.015 | 0.013 | 0.126 | 0.107 | 0.021 | 0.016 | ||
| YFP | 233.88 | 229.97 | 199.54 | 193.24 | 57.31 | 58.69 | 171.68 | 148.36 | 60.58 | 63.76 | ||
| YFP/OD | 1016.869565 | 1017.566372 | 997.7 | 971.0552764 | 3820.666667 | 4514.615385 | 1362.539683 | 1386.542056 | 2884.761905 | 3985 | ||
| (-) Ctrl (P41) | A | Dilution | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | N/A | N/A | N/A | N/A | 1 in 10 | 1 in 10 |
| OD | 0.232 | 0.215 | 0.197 | 0.199 | 0.016 | 0.015 | 0.111 | 0.133 | 0.019 | 0.018 | ||
| YFP | 227.67 | 216.53 | 186.19 | 198.19 | 65.69 | 65.34 | 150.96 | 162.94 | 60.46 | 59.28 | ||
| YFP/OD | 981.3362069 | 1007.116279 | 945.1269036 | 995.9296482 | 4105.625 | 4356 | 1360 | 1225.112782 | 3182.105263 | 3293.333333 | ||
| B | Dilution | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | N/A | N/A | N/A | N/A | 1 in 20 | 1 in 10 | |
| OD | 0.227 | 0.234 | 0.199 | 0.197 | 0.029 | 0.027 | 0.228 | 0.125 | 0.021 | 0.022 | ||
| YFP | 237.44 | 226.99 | 190.07 | 186.35 | 71.71 | 71.9 | 241.84 | 160.57 | 59.6 | 61.7 | ||
| YFP/OD | 1045.991189 | 970.042735 | 955.1256281 | 945.9390863 | 2472.758621 | 2662.962963 | 1060.701754 | 1284.56 | 2838.095238 | 2804.545455 | ||
| C | Dilution | 1 in 4 | 1 in 4 | 1 in 4.5 | 1 in 4.5 | N/A | N/A | N/A | N/A | 1 in 10 | 1 in 10 | |
| OD | 0.219 | 0.218 | 0.193 | 0.194 | 0.017 | 0.018 | 0.134 | 0.118 | 0.021 | 0.019 | ||
| YFP | 224.51 | 227.38 | 201.61 | 190.81 | 59.56 | 61.47 | 167.86 | 162.14 | 59.57 | 57.91 | ||
| YFP/OD | 1025.159817 | 1043.027523 | 1044.611399 | 983.556701 | 3503.529412 | 3415 | 1252.686567 | 1374.067797 | 2836.666667 | 3047.894737 |
Sequencing Parts, Plasmids, Etc.
Primers for Sequencing Remy's Plasmids and Duet Vectors 7/21
| Primer Name | Sequence |
| P4P13fwd | GGATCTCGACGCTCTCCCTT |
| P12fwd | ATGCGTCCGGCGTAGAGGA |
| DuetRev | TGCTAGTTATTGCTCAGCGG |
These arrived 7/25/08, and were reconstituted to 100uM.
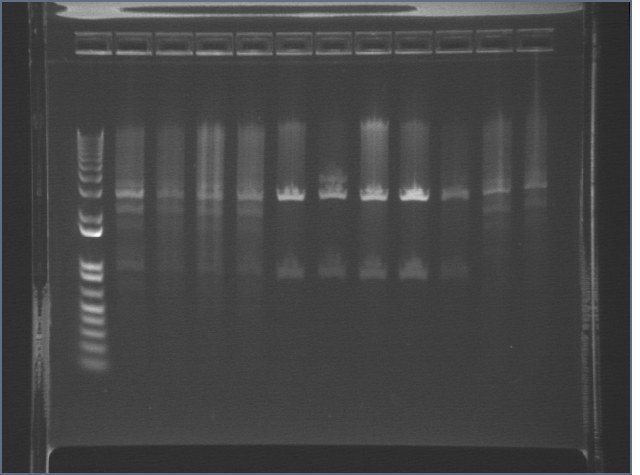
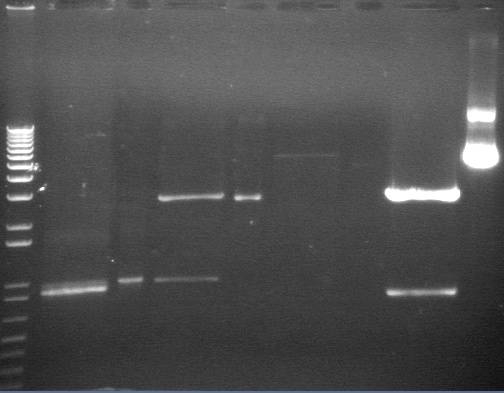
Analytical Digest of Composite Parts 7/22
Digested composite plasmids to verify size.
| Plasmid | DNA Added (uL) | 10x Buffer 3 (uL) | 100x BSA (uL) | XbaI (uL) | PstI (uL) | Water (uL) | Total (uL) | < 1 ug DNA? | Expected Size |
| S1 P75a 1 (7/22) | 7 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 13.25 | 25 uL | 925bp, 2750bp | |
| S1 P75a 2 (7/22) | 5 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 15.25 | 25 uL | 925bp, 2750bp | |
| S1 P75b 1 (7/22) | 7 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 13.25 | 25 uL | 925bp, 2750bp | |
| S1 P75b 2 (7/22) | 4 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 16.25 | 25 uL | 925bp, 2750bp | |
| E1 P92a 1:1 (7/17) | 7 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 13.25 | 25 uL | 2314bp, 2750bp | |
| E1 P92b 1:1 (7/17) | 7 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 13.25 | 25 uL | 2314bp, 2750bp | |
| E1 P93a 1:1 (7/17) | 7 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 13.25 | 25 uL | 2314bp, 2750bp | |
| E1 P93b 1:1 (7/17) | 10 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 10.25 | 25 uL | 2314bp, 2750bp | |
| E1 P92a PCR (7/18) | 15 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 5.25 | 25 uL | Yes | 2314bp, 2750bp |
| S1 P93 1:3 1 (7/17) | 7 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 13.25 | 25 uL | 2314bp, 2750bp | |
| S1 P93 1:3 2 (7/17) | 7 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 13.25 | 25 uL | 2314bp, 2750bp | |
| S1 P92a PCR 1 (7/18) | 7 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 13.25 | 25 uL | 2314bp, 2750bp | |
| S1 P92a PCR 2 (7/18) | 7 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 13.25 | 25 uL | 2314bp, 2750bp | |
| S1 P93b PCR 1 (7/18) | 4 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 16.25 | 25 uL | 2314bp, 2750bp | |
| S1 P93b PCR 2 (7/18) | 7 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 13.25 | 25 uL | 2314bp, 2750bp | |
| E1 P58b 1 (7/18) | 7 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 13.25 | 25 uL | 937bp, 2750bp | |
| E1 P58b 2 (7/18) | 10 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 10.25 | 25 uL | 937bp, 2750bp | |
| S1 P58b A (7/15) | 10 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 10.25 | 25 uL | Yes | 937bp, 2750bp |
| S1 P58b B (7/15) | 10 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 10.25 | 25 uL | Yes | 937bp, 2750bp |
| S1 P59 (7/9) | 10 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 10.25 | 25 uL | Yes | 1122bp, 2750bp |
| S1 P59b 1 (7/22) | 10 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 10.25 | 25 uL | Yes | 1122bp, 2750bp |
| S1 P59b 2 (7/18) | 3 | 2.5 uL | 0.25 uL | 1 uL | 1 uL | 17.25 | 25 uL | 1122bp, 2750bp |
Gel Results 7/23
Making Thermoinducible cI Lambda System
Primers for Taking Out cI857 from PGW7 7/21
| Primer Name | Sequence |
| CI857_RBSfwd | Atcgagaattcgcggccgcttctagagaaagaggagaaaatgagcacaaaaaagaaac |
| CI857_RBSrev | Atcgatactagtagcggccgctgcagtcagccaaacgtctcttcag |
| CI857_BIGfwd | Atcgagaattcgcggccgcttctagagtcatgacattaacctataaa |
| CI857_BIGrev | atcgatactagtagcggccgctgcagcagccagcagagaattaagg |
Minipreps of Transformed Parts in S1 7/22
Also includes transformed P28 and the thermoinducible lac system.
| Plasmid Name | ng/uL | A260 | 260/280 | 260/230 | Constant |
| S1 P75a 1 | 156.2 | 3.124 | 1.99 | 2.27 | 50 |
| S1 P75a 2 | 203.24 | 4.065 | 2.04 | 2.23 | 50 |
| S1 P75b 1 | 177.41 | 3.548 | 2.02 | 2.16 | 50 |
| S1 P75b 2 | 258.85 | 5.177 | 2.01 | 2.3 | 50 |
| S1 P28a 1 | 155.42 | 3.108 | 2.03 | 2.27 | 50 |
| S1 P28a 2 | 239.51 | 4.79 | 2.06 | 2.26 | 50 |
| S1 P28a 3 | 204.5 | 4.09 | 2.01 | 2.24 | 50 |
| S1 P93 1:3 1 | 175.27 | 3.505 | 2.07 | 2.32 | 50 |
| S1 P93 1:3 2 | 166.9 | 3.338 | 2.12 | 2.38 | 50 |
| S1 P92a PCR 1 | 152.58 | 3.052 | 2.02 | 2.28 | 50 |
| S1 P92a PCR 2 | 157.59 | 3.152 | 2.02 | 2.22 | 50 |
| S1 P93b PCR 1 | 291.87 | 5.837 | 2.13 | 2.23 | 50 |
| S1 P93b PCR 2 | 152 | 3.04 | 2.07 | 2.21 | 50 |
Adding Terminator Before pLambda GFP
Digestion of Terminator (P64) and pLambda GFP 7/22
| ' | P75a 1 (vector) | P75b 1 (vector) | P64a 07 (insert) |
| DNA | 10 uL | 10 uL | 10 uL |
| 100x BSA | 0.25 uL | 0.25 uL | 0.25 uL |
| 10x Buffer | 2.5 uL EcoRI buffer | 2.5 uL EcoRI buffer | 2.5 uL EcoRI buffer |
| Enzyme 1 | 1 uL EcoRI | 1 uL EcoRI | 1 uL EcoRI |
| Enzyme 2 | 1 uL XbaI | 1 uL XbaI | 1 uL SpeI |
| Water | 5.25 uL | 5.25 uL | 5.25 uL |
| Total | 25 uL | 25 uL | 25 uL |
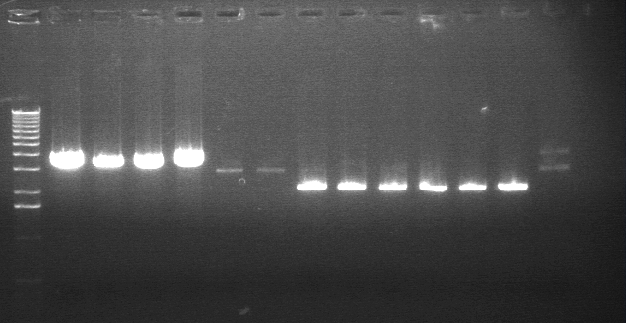
Digestion Gel Results 7/23
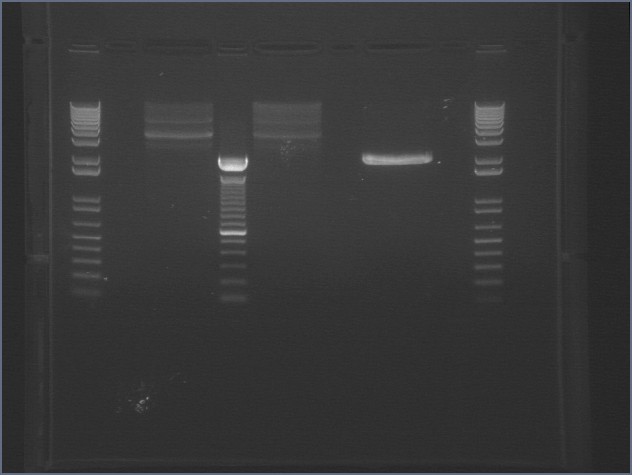
| 1% Agarose, visualized using EtBr/UV | |
|---|---|---|
| Lane | Sample | |
| 1 | 1kB Ladder | |
| 3 | S1 P75a (2750/925) | |
| 4 | 100bp Ladder | |
| 5 | S1 P75b 1 (2750/925) | |
| 7 | P64a 07 (~3kb/129bp) | |
| 9 | 1kB Ladder | |
Extracted and purified S1 P75a 1 and P75b 1, but P64 07 has incorrect bands.
Desphosphorylated S1 P75a 1 and P75b 1 same day.
Digestion of (Almost) All of Our Terminators 7/23
| ' | P41 | P61 | P62a 07 | P64 |
| DNA | 5 uL | 8 uL | 8 uL | 8 uL |
| 100x BSA | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL |
| 10x EcoRI Buffer | 2.5 uL | 2.5 uL | 2.5 uL | 2.5 uL |
| Enzyme 1 | 1 uL EcoRI | 1 uL EcoRI | 1 uL EcoRI | 1 uL EcoRI |
| Enzyme 2 | 1 uL SpeI | 1 uL SpeI | 1 uL SpeI | 1 uL SpeI |
| Water | 15.25 uL | 15.25 uL | 15.25 uL | 15.25 uL |
| Total | 25 uL | 25 uL | 25 uL | 25 uL |
Gel? 7/24
Faint band was detected in P64 (?) but was not visible under UV when cutting, so not extracted. Will try digestion of P63, which has worked for Meng Xiao and Thilini before.
Ligation of P75 with old P63 7/24
Purified P63 cut with ES was ligated with the dephosphorylated P75 vector. Very little of the insert was available (~2 uL)
Transformed on Kan plates.
7/25: Almost all plates were blank except for positive control. One faint, questionable colony found on plate with 1 uL ligation reaction-- picked for Saturday.
Digestion of P63 (7/24) and E1 P75b (7/25)
| ' | P63a | P63b | E1 P75b |
| DNA | ? | ? | 10 uL |
| 100x BSA | 0.25 uL | 0.25 uL | 0.25 uL |
| 10x EcoRI Buffer | 2.5 uL | 2.5 uL | 2.5 uL |
| Enzyme 1 | 1 uL EcoRI | 1 uL EcoRI | 1 uL EcoRI |
| Enzyme 2 | 1 uL SpeI | 1 uL SpeI | 1 uL XbaI |
| Water | ? | ? | 10.25 uL |
| Total | 25 uL | 25 uL | 25 uL |
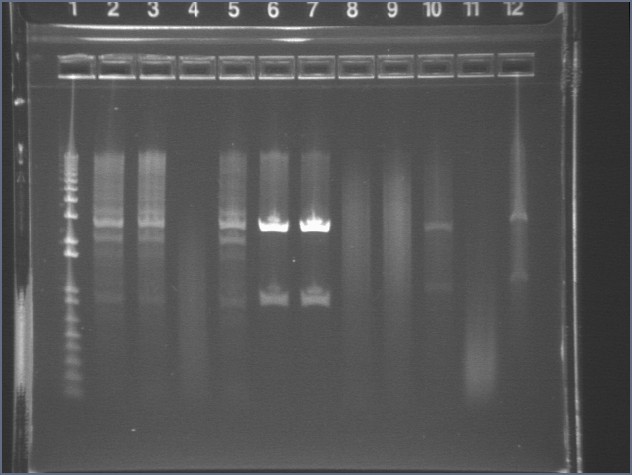
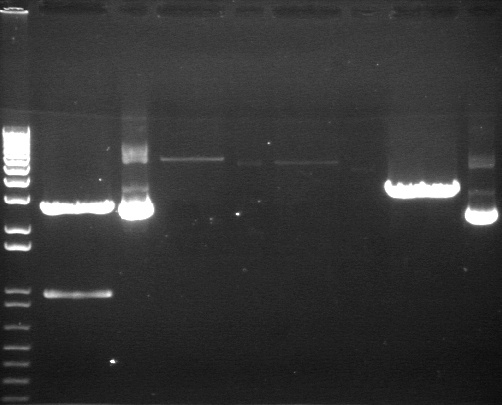
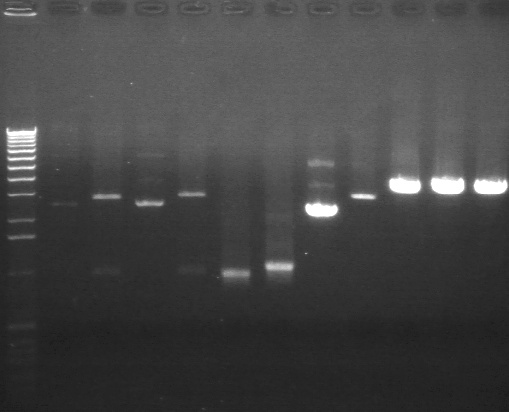
Gel Results 7/25
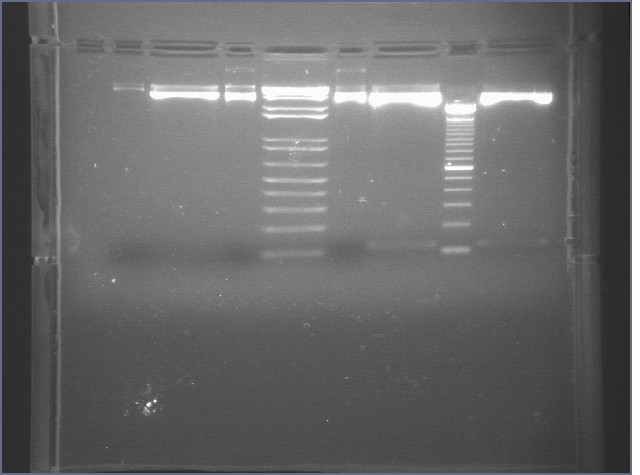
| 1% Agarose, visualized using EtBr/UV | |
|---|---|---|
| Lane | Sample | |
| 3 | E1 P75b (2750/ 925 bp) | |
| 5 | 1 kb Ladder | |
| 7 | P63a (95 bp) | |
| 8 | 100 bp Ladder | |
| 9 | P63b (95 bp) | |
95 bp bands became fainter (and undetectable with gel mold on) after running the gel for more time.
E1 P75b, P63a, and P63b were extracted and purified. E1 P75b was dephosphorylated using the Roche T4 Ligation Kit Alkaline Phosphatase.
Ligation of pLambda+GFP Vector P75 with Terminator P63 7/25
| ' | E1 P75b + P63a (6:2) | E1 P75b + P63b (6:2) | S1 P75a + P63a (6:2) | S1 P75a + P63b (6:2) | S1 P75b + P63a (6:2) | S1 P75b + P63b (6:2) | E1 P75b + P63a (5:1) | E1 P75b + P63b (5:1) | S1 P75a + P63a (5:1) | S1 P75b + P63b (5:1) |
| Insert | 6 uL | 6 uL | 6 uL | 6 uL | 6 uL | 6 uL | 5 uL | 5 uL | 5 uL | 5 uL |
| Vector | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL | 1 uL | 1 uL | 1 uL | 1 uL |
| DNA Dilution Buffer | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL |
| Water (to 20 uL) | 0 uL | 0 uL | 0 uL | 0 uL | 0 uL | 0 uL | 2 uL | 2 uL | 2 uL | 2 uL |
| Ligation Buffer | 10 uL | 10 uL | 10 uL | 10 uL | 10 uL | 10 uL | 10 uL | 10 uL | 10 uL | 10 uL |
| Ligase | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL |
Transformation of P75+ P63 Ligations and PGW7
| Plate | Marker | # Colonies | Plate Description |
| E1 P75b + P63a (6:2) | Kan | 0 | |
| E1 P75b + P63b (6:2) | Kan | 0 | |
| S1 P75a + P63a (6:2) | Kan | 1 | Single colony is fluorescent. |
| S1 P75a + P63b (6:2) | Kan | 0 | |
| S1 P75b + P63a (6:2) | Kan | 1 | Single colony NOT fluorescent. |
| S1 P75b + P63b (6:2) | Kan | 0 | |
| E1 P75b + P63a (5:1) | Kan | 0 | |
| E1 P75b + P63b (5:1) | Kan | 0 | |
| S1 P75a + P63a (5:1) | Kan | 0 | |
| S1 P75b + P63b (5:1) | Kan | 0 | |
| PGW7 1 (1 uL plasmid in 50 uL cells) | Amp | 200+ | |
| PGW7 2 (1 uL plasmid in 100 uL cells) | Amp | 200+ | |
| puc19 30s Heatshock | Amp | 18 | |
| puc19 45s Heatshock | Amp | 15 | |
| puc19 1m Heatshock | Amp | 6 | |
| puc19 2m Heatshock | Amp | 14 | |
| Kan (-) ctrl | Kan | 0 | |
| Amp (-) ctrl | Amp | 0 |
Gradient PCR of PGW7 Plasmid to Isolate cI857 7/25
Used primer sets RBS and BIG. Prepared enough for 4 standard reactions (50 uL each), and split into 8 reactions in strip top tubes.
Reaction Recipe:
| Fwd Primer | 1 uL |
| Rev Primer | 1 uL |
| Platinum Supermix | 45 uL |
| Water | 2 uL |
Added a total of 2 uL of PGW7 plasmid to each reaction mix (0.25 uL plasmid per 25 uL reaction).
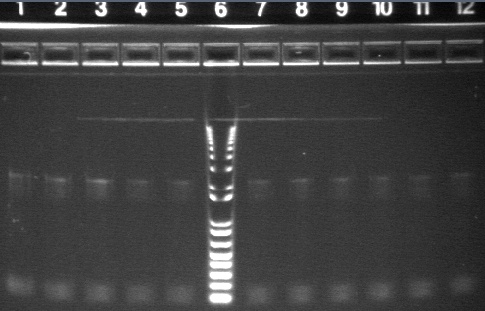
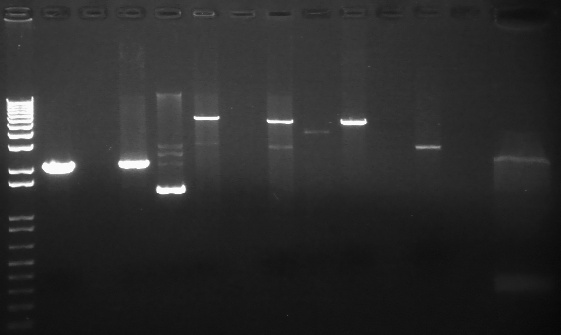
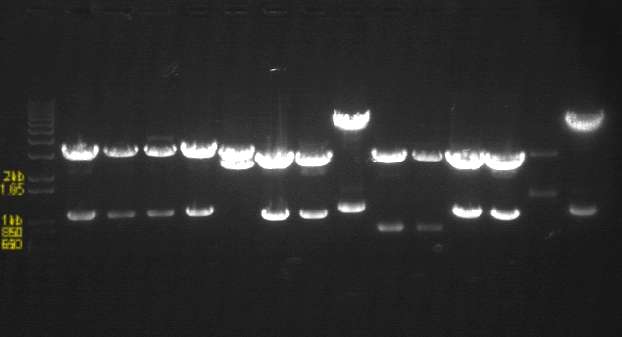
PCR Gels 7/25
PCR using RBS Primer Set:
- Amplified region is cI857 coding region only, and primers have Biobricks prefix, suffix, and RBS.
- 675bp
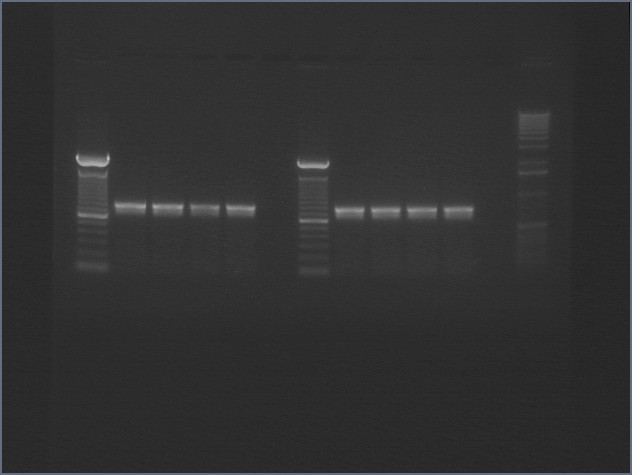
| 1% Agarose, visualized using EtBr/UV | |
|---|---|---|
| Lane | Annealing Temperature | |
| 1 | 100 bp Ladder | |
| 2 | 40.8 | |
| 3 | 41.5 | |
| 4 | 42.5 | |
| 5 | 43.9 | |
| 7 | 100bp Ladder | |
| 8 | 45.4 | |
| 9 | 46.7 | |
| 10 | 47.6 | |
| 11 | 48.3 | |
| 13 | 1 kB Ladder | |
All bands were extracted and purified.
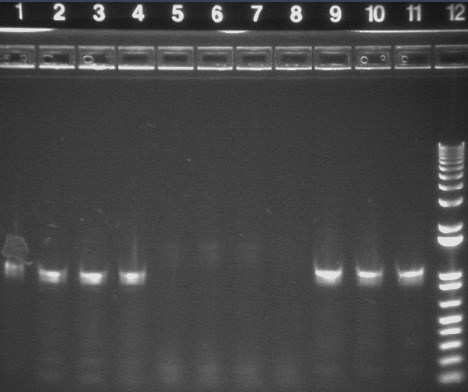
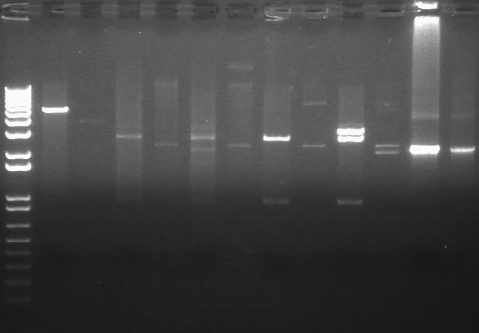
PCR using BIG Primer Set:
- Amplified region is cI857 coding region and 200bp upstream and downstream, and primers have Biobricks prefix and suffix.
- 1193bp
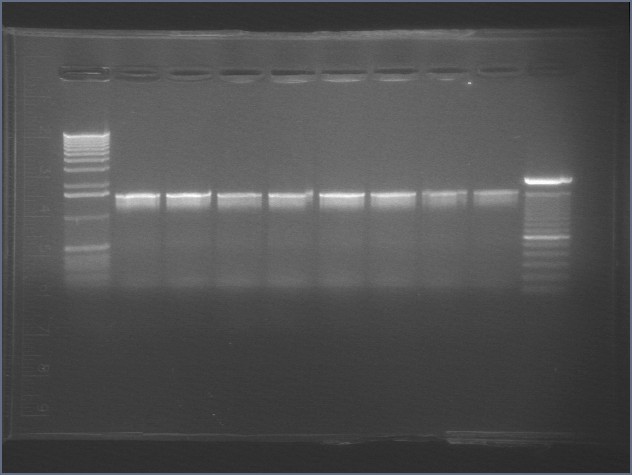
| 1% Agarose, visualized using EtBr/UV | |
|---|---|---|
| Lane | Annealing Temperature | |
| 1 | 1 kB Ladder | |
| 2 | 40.8 | |
| 3 | 41.5 | |
| 4 | 42.5 | |
| 5 | 43.9 | |
| 6 | 45.4 | |
| 7 | 46.7 | |
| 8 | 47.6 | |
| 9 | 48.3 | |
| 10 | 100 bp Ladder | |
All bands were extracted and purified.
Housekeeping
Making Competent Cells 7/22
Made ~600 100uL (some were 200 uL, indicated by green dot on top) aliquots from 1L of culture. Done as per Jason's lab's protocol.
New cells seem to work, but tested heatshock times with puc19 7/25/08.
Ligations 07/22
- Attempted to ligate p40 cut with SP (w/ RBS) to mtrB cut with XP. Thus far, the ligation has not worked.
Recombination Update 07/22
- Thus far, none of the attempts to extract flanking regions from the genomic DNA have worked. We are troubleshooting this now and will run positive controls with known DNA samples to check the efficiency of our Phusion Polimerase.
07/23 Ligations/Transformations
- Ligated p40 cut SP with mtrB cut XP at 2:3, 1:2, 1:4, 1:5, 1:6, positive control is uncut p40, negative control (to test Amp plates) is p29 (Kan)
Ligations 07/24
We ligated p75a w/ p63 w/ a ratio of 4ul p63:1ul p75a
Transformed into new dh5α competent cells w/ volumes of 1, 3, and 5uL, using Jason's new protocol. As a control transformed 1uL of pUC19.
PCR
mtrB
7/22: gradient + new R primer
Rx mix (split into 12 samples): 180μL PCR supermix, 4μL mtrB-ApaLI-F primer (20μM), 4μL mtrB-KpnI-R-new (20μM), pipet tip touch of mtrB BB from gel purification, 12μL water Rx: 5min @ 94°C → 35x[45s @ 94°C → 45s @ {52-58 gradient}°C → 2m38s @72°C] → 5min @ 72°C → ∞ @ 4°C
No bands on gel from any of the conditions.
Lower gradient
Rx mix (split into 12 samples): 180μL PCR supermix, 4μL mtrB-ApaLI-F primer (20μM), 4μL mtrB-KpnI-R-new (20μM), pipet tip touch of WT S. oneidensis MR-1, 12μL water Rx: 10min @ 94°C → 40x[45s @ 94°C → 45s @ {44-51 gradient}°C → 2m45s @72°C] → 5min @ 72°C → ∞ @ 4°C
7/23: gel
All the temperatures seem to have worked (51 was not loaded due to # lanes on gel). Expected product size ~2.1kb. Ladder is 1KB.
Products from different temperatures combined and gel purified.
P51, 52, 17 MIT
17, 52:
Rx mix (split into 4 samples): 180μL Platinum PCR supermix, 4μL BBpfx primer (20μM), 4μL BBsfx primer (20μM), 4μL miniprepped DNA, 8μL water
51:
Rx mix (split into 4 samples): 100μL AmpliTaq Gold Master Mix, 4μL BBpfx primer (20μM), 4μL BBsfx primer (20μM), 4μL miniprepped DNA, 88μL water
Rx: 5min @ 94°C → 35x[45s @ 94°C → 45s @ {52.7, 53.6, 54.6, 55.5}°C → 1m25s @72°C] → 5min @ 72°C → ∞ @ 4°C
Results
P1, P3 backbones; HO-pcyA; P51 MIT
Conditions optimization
Rx: 5min @ 94°C → 35x[45s @ 94°C → 45s @ {middle 8 of 41-55 gradient: 43.3, 44.9, 47.0, 49.3, 51.3, 52.8, 53.9, 54.7}°C → 3m30s @72°C] → 5min @ 72°C → ∞ @ 4°C
P1, P3
PCR product is the backbone of P1 and P3, including the RBS (B0032), weak and strong promoter respectively, and terminator. The ends have unique ApaLI and KpnI cut sites.
Rx mix (split into 8 samples): 180μL Platinum PCR supermix, 4μL P3minusGFP-F primer (20μM), 4μL P3minusGFP-R primer (20μM), 4μL miniprepped DNA (P1/P3), 8μL water
HO-pcyA
Ends have unique ApaLI and KpnI cut sites.
Rx mix (split into 8 samples): 180μL Platinum PCR supermix, 4μL HO-pcyA-F primer (20μM), 4μL HO-pcyA-R primer (20μM), 4μL miniprepped P86, 8μL water
P51 MIT
Rx mix (split into 4 samples): 90μL Platinum PCR supermix, 2μL BBsfx primer (20μM), 2μL BBpfx primer (20μM), 2μL miniprepped P51, 4μL water
Used highest 4 temps of gradient.
Gels
Annealing temp increases LTR.
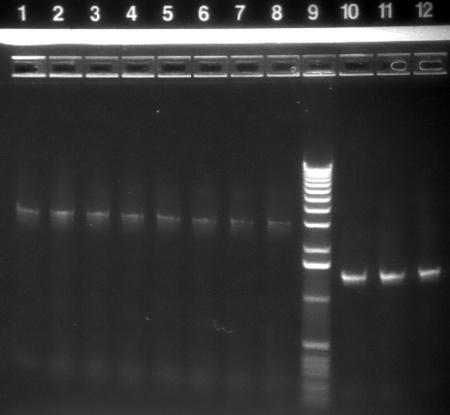
| 1.2% agarose E-gel run for 30 min and visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents | |
| 1-8 | P1 backbone (~3kb) | |
| 9 | 1 KB ladder | |
| 10-12 | HO-pcyA (~1.4 kb) | |

| 1.2% agarose E-gel run for 30 min and visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents | |
| 1-5 | HO-pcyA (~1.4 kb) | |
| 6 | 1 KB ladder | |
| 7-12 | P3 backbone (~3 kb) | |
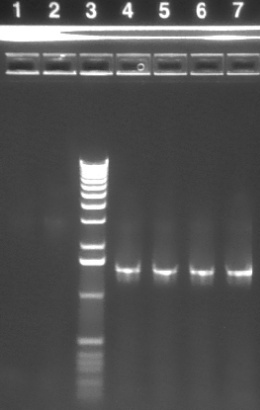
| 1.2% agarose E-gel run for 30 min and visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents | |
| 1-2 | P3 backbone (~3kb) | |
| 3 | 1 KB ladder | |
| 4-7 | P51 MIT (~1.4 kb) | |
Full Rx
400 μL (8 reactions) each of P1 and P3 were set up using a doubling of above reaction mix. The reaction conditions were the same except that annealing temperature was 45°C, and cycles occur 40 times.
100 μL (2 reactions) each of HO-pcyA and P51 MIT were set up using above reaction ratios. 40 cycles were performed with 55°C annealing temp and 1:35 extension time.
Products will be gel purified along with products from optimization PCRs.
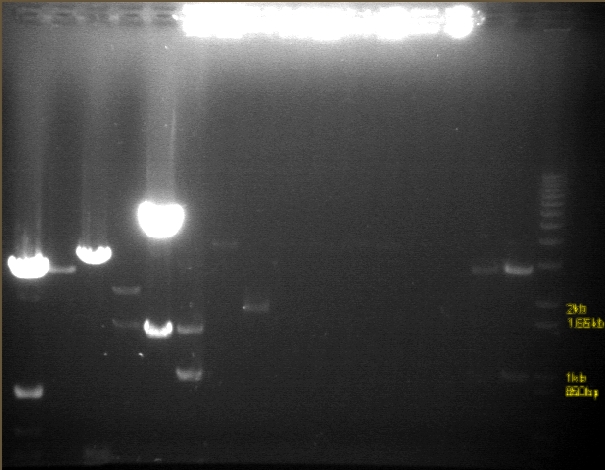
RE digests 07/22
Gel 1
Gel 2
Gel 3

| 1 agarose gel visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents | |
| 1 | 1 kb plus ladder | |
| 2 | P97A cut EX (2091) | |
| 3 | P97A uncut (2091) | |
| 4 | P97B cut EX (2091) | |
| 5 | P97B uncut (2091) | |
Ligation w/ purified products 7/22: CDF into P1 vector
- We ligated P1A (vector) to CDF. We used three different ratios of vector to insert for the ligation: 2/6, 1/5, 1/7.
RE digests 07/23
Ligations with purified digest fragments 07/23: mtrB+RBS, CDF into pSB3K3
We ligated
- P97 and mtrB BB
- P3 and CDF
We used three different ratios of insert to vector for the ligation: 6/2, 5/1, 7/1.
We used 5 μL of the ligation to transform TOP10 cells and 5 μL to transform the new DH5α cells. We also used 1 μL and 3 μL to transform DH5α. We used pUC19 (1 μL) as a positive control to test the efficacy of the new DH5α cells.
We used Jason's protocol to transform the new DH5α cells:
- Thaw cells by resting tubes on ice
- Then add DNA and mix by swirling with the pipette tip
- Incubate the cells with DNA in ice for 10min
- Heat shock at 42C for 2min
- Then incubate in ice for 2-5min
- Add 500-700ml LB (or SOC) and incubate + 250rpm shaking for 1hr at 37C
- Plate
Results
| Strain | DNA | Vector:Insert (μL) | Amount transformed (μL) | Plate | # colonies |
| E1 | pUC19 | n/a | 1 | AMP | 40 |
| E1 | - | n/a | - | CARB | 0 |
| E2 | P97+ mtrB BioBrick | 2:6 | 5 | CARB | 136 |
| E2 | P97+ mtrB BioBrick | 1:5 | 5 | CARB | 48 |
| E2 | P97+ mtrB BioBrick | 1:7 | 5 | CARB | 64 |
| E1 | P97+ mtrB BioBrick | 1:7 | 1 | CARB | 0 |
| E1 | P97+ mtrB BioBrick | 1:7 | 3 | CARB | 0 |
| E1 | P97+ mtrB BioBrick | 1:7 | 5 | CARB | 4 |
| E1 | P97+ mtrB BioBrick | 1:5 | 1 | CARB | 0 |
| E1 | P97+ mtrB BioBrick | 1:5 | 1 | CARB | 0 |
| E1 | P97+ mtrB BioBrick | 1:5 | 1 | CARB | 0 |
| E1 | P97+ mtrB BioBrick | 2:6 | 1 | CARB | 1 |
| E1 | P97+ mtrB BioBrick | 2:6 | 1 | CARB | 7 |
| E1 | P97+ mtrB BioBrick | 2:6 | 1 | CARB | 9 |
| E2 | P3+CDF | 2:6 | 5 | KAN | 7 |
| E2 | P3+CDF | 1:5 | 5 | KAN | 56 |
| E2 | P3+CDF | 1:7 | 5 | KAN | 5 |
| E1 | P3+CDF | 1:7 | 1 | KAN | 0 |
| E1 | P3+CDF | 1:7 | 3 | KAN | 2 |
| E1 | P3+CDF | 1:7 | 5 | KAN | 0 |
| E1 | P3+CDF | 1:5 | 1 | KAN | 0 |
| E1 | P3+CDF | 1:5 | 3 | KAN | 0 |
| E1 | P3+CDF | 1:5 | 5 | KAN | 0 |
| E1 | P3+CDF | 2:6 | 1 | KAN | 0 |
| E1 | P3+CDF | 2:6 | 3 | KAN | 1 |
| E1 | P3+CDF | 2:6 | 5 | KAN | 0 |
- Try 45s heat shock and SOC for new cells
- Try 3 or 5 μL transformations
- New Carb plates seem fine
RE digests of miniprepped ligations 07/25
Lane 1: 1 kb ladder
Lanes 2-7: P90 A-F cut SP (~3600)
Lanes 8-13: P98 A-F cut ES (~2100)
Lane 14: P3 cut XP (2750)
Ligations with purified parts and PCR products 07/26
Ligations were performed using standard protocol. New homemade cells were heat shocked for 40s, and incubated in 200μL SOC for 1 hour prior to plating on prewarmed plates. Vectors were dephosphorylated with Antarctic Phosphatase as per NEB's protocol.
Results 7/27
| DNA | Strain | Vector,Insert | Plate | No. of Colonies |
| P3+HO-pcyA not BB PCR | E1 | 3,5 | Kan | 1 |
| P3+HO-pcyA not BB PCR | E2 | 2,6 | Kan | ~50 |
| P1+HO-pcyA not BB PCR | E2 | 2,6 | Kan | ~50 |
| P1+HO-pcyA not BB PCR | E1 | 3,5 | Kan | 0 |
| P1+mtrB not BB PCR | E2 | 2,6 | Kan | 32 |
| P1+mtrB not BB PCR | E1 | 3,5 | Kan | 0 |
| P3+mtrB not BB PCR | E1 | 3,5 | Kan | 13 |
| P3+mtrB not BB PCR | E2 | 2,6 | Kan | ~80 |
| P63+98 | E1 | 1,7 | Amp | TMTC |
| P63+98 | E2 | 2,6 | Amp | TMTC |
| P3+51 PCR | E2 | 2,6 | Kan | ~120 |
| P3+51 PCR | E1 | 3,5 | Kan | 59 |
| P90+49 | E1 | 1.5,6.5 | Amp | TMTC |
| P90+49 | E2 | 2,6 | Amp | TMTC |
| P90+48 | E1 | 1.5,6.5 | Cm | ~80 |
| P90+48 | E2 | 2,6 | Cm | TMTC |
| pUC19 (positive control) | E1 | 1 ul | Carb | ~200 |
2 non-fluorescent colonies picked for 5mL cultures for each plate when possible.
RE digests 07/24
Ligations w/ purified digest fragments 07/24
Vector:insert ratio: 2:6 μL, volume used in transformation: 4μL
Note that 45s heat shock was used for DH5α, and cells were split onto 2 plates to accelerate drying.
Results 7/25
| DNA | Strain | Plate | # Colonies |
| P1+P17 | E2 | KAN | 184 |
| P1+P17 | E1 | KAN | 9 |
| P1+P17 | E1 | KAN | 9 |
| P1+P52 | E2 | KAN | 40 |
| P1+P52 | E1 | KAN | 0 |
| P1+P52 | E1 | KAN | 0 |
| P3+P17 | E2 | KAN | 10 |
| P3+P17 | E1 | KAN | 0 |
| P3+P17 | E1 | KAN | 0 |
| P3+P52 | E2 | KAN | 48 |
| P3+P52 | E1 | KAN | 1 |
| P3+P52 | E1 | KAN | 1 |
| P38+P17 | E2 | KAN | 1 |
| P38+P17 | E1 | KAN | 0 |
| P38+P17 | E1 | KAN | 0 |
| P38+P51 | E2 | KAN | 32 |
| P38+P51 | E1 | KAN | 1 |
| P38+P51 | E1 | KAN | 0 |
| P38+P52 | E2 | KAN | 1 |
| P38+P52 | E1 | KAN | 0 |
| P38+P63 | E1 | KAN | 0 |
| P39+P17 | E1 | KAN | 0 |
| P39+P17 | E1 | KAN | 0 |
| P39+P17 | E2 | KAN | 0 |
| P39+P51 | E2 | KAN | 5 |
| P39+P51 | E1 | KAN | 2 |
| P39+P51 | E1 | KAN | 2 |
| P39+P52 | E2 | KAN | 6 |
| P39+P52 | E1 | KAN | 1 |
| P39+P52 | E1 | KAN | 1 |
| P90+P48 | E2 | CM | 16 |
| P90+P48 | E1 | CM | 5 |
| P90+P48 | E1 | CM | 1 |
| P90+P49 | E2 | AMP | TMTC |
| P90+P49 | E1 | AMP | 19 |
| P90+P49 | E1 | AMP | 4 |
| pUC19 | E1 | AMP | 192 |
Cultures for miniprep
When possible, 2 non-fluorescent colonies were picked from each plate (fluorescence in the P1/P3 derived plasmids indicates P1/P3 vector religation).
- The 2 E2 P90+49 cultures failed to grow. Note that Kan selection was used, as that's carried on P90, although the transformation plates were Amp.
RE digests of minipreps 07/26
We digested
- P46 with NheI
- P90+49 and P90+48 with XbaI and SpeI
- When these parts are combined, we will have a plasmid with the CDF origin (CDF+Amp and CDF+Cm in the base vector)
- P3+17 and P1+17 with EcoRI and XbaI
- We can add the high and low constitutive promoters to give us the complete Tet system (without GFP) in a p15A vector
- P39+51, P38+51, P38+17 with XbaI and PstI
- We can put these parts into P1 and P3 to give us the complete Tet and Lac systems in a p15A vector
Results
Cultures were set up again for samples 7-14, since the DNA seems to have not run properly.
 "
"