Team:Heidelberg/Notebook/Killing II/4thweek
From 2008.igem.org


4th week
Contents |
Monday 08/25/2008
Colicin Receiver
pSB1A3-Receiver-Colicin-Cloning
- PCR of colicin E1 and E9 operon with specific primers:
25.0 µl Phusion MasterMix 2.5 µl Primer fw(ColE1_prot_fw_BamHI/ColE9_plasmid_rv_SpeI) 2.5 µl Primer rv(ColE1_kil_prot_rv_SpeI/ColE9_plasmid_rv_SpeI) 17.0 µl H2O 3.0 µl pColE1/pColE9-J ------- 50.0 µl
program: 98 °C 30 sec 98 °C 10 sec | 57 °C 20 sec | 25 cycles 72 °C 45 sec | 72 °C 8 min 4 °C constant
Activity Test
- Inoculation of 5 ml liquid ONC with...
- pColE1 in JC411 -> 0.1% Glucose Medium
- pColE9 in MG1655
- TOP 10
- MG 1655
Sender part
pBAD-Sender Cloning
- Controldigestion of BBa_F1610 with DraI: 1.5 - 2 h -> 37 °C
10 µl DNA (320-330 ng/µl) 3 µl Tango Buffer 10x (Fermentas) 2 µl DraI (Fermentas) 15 µl H2O ----- 30 µl
- Gel of digestion: Results are not perfect but expected bands can be estimated.
Activity Test
- Inoculation of pBAD-BBa_F1610 in 8 ml M9 Kana + 0.1% Arabinose. (8 colonies)
General
- Inoculation of liquid ONC of BBa_J23107, BBa_J23102, BBa_R0011, BBa_R0040 in 5 ml LB Amp. (For Minipreps and Glycerolstocks)
[back]
Tuesday 08/26/2008
Colicin Receiver
pSB1A3-Receiver-Colicin-Cloning
- PCR Purification of T9002 without GFP (08/22/08) and colicin E1/E9 operon: Qiagen PCR Purification Kit.
- Digestion of purified products with BamHI: 2h -> 37 °C
20.0 µl DNA (PCR product, purified) 13.0 µl H2O 4.0 µl NEBuffer 3 (NEB) 3.0 µl BamHI (NEB) 4.0 µl BSA 10x (NEB) ------- 44.0 µl
- Gelextraction of digestion: Cutted the bands and freezed for purification tomorrow.
Activity Test
- 2nd step of colicin test based on ONC from Tuesdaysday:
- pColE1 (JC411):
- inoculation of 12 ml LB ONC + 0.1% Glucose -> unstressed
- inoculation of 12 ml LB ONC + 0.1% Glucose + 1µg/ml Mytomycin C
- EDIT 09/02/08: Too high Mytomycin C concentrations
- 100 µl ONC + 300 µl LB + 200 µl 10% glucose plated on LB-Agar
- pColE9 (MG1655):
- inoculation of 12 ml LB ONC-> unstressed
- inoculation of 12 ml LB ONC + 1µg/ml Mytomycin C
- EDIT 09/02/08: Too high Mytomycin C concentrations
- 100 µl ONC + 500 µl LB plated on LB-Agar
- MG1655 + TOP10:
- inoculation of 12 ml LB ONC-> unstressed
- 100 µl ONC + 500 µl LB plated on LB-Agar
- pColE1 (JC411):
Sender part
pBAD-Sender Cloning
- Control of Cloning -> see activity test
Activity Test
- To check if we have one positive clone we perform the sender activity test again (see Fridayday 08/08/22 for plate scheme and details). After the first measurements an increase in GFP expression by BBa_T9002 was observed for colonies 9, 10, 11. Because of that we inoculated liquid ONC with LB-Kana for miniprepes and glycerolstocks
[back]
Wednesday 08/27/2008
Colicin Receiver
pSB1A3-Receiver-Colicin-Cloning
- Gelextraction of BamHI digestion: Qiagen Gelextraction kit
- Ligation of receiver with colicins ratio 1:1: 1h -> RT, 10 min -> 65 °C
10 µl DNA I 10 µl DNA II 4 µl 10x T4 DNA Ligase Buffer 2 µl T4 Ligase 14 µl H2O ----- 40 µl
- Gelextaction of Ligation: ligated construct (~3000 bp) was extracted and purified. Qiagen Gel Extraction Kit
- Digestion of ligated product with XbaI and SpeI: 1.5 h -> 37 °C and 65 °C -> 20 min
20 µl DNA 6 µl H2O 4 µl NEBuffer 2 3 µl SpeI 3 µl XbaI 4 µl BSA 10x ----- 40 µl
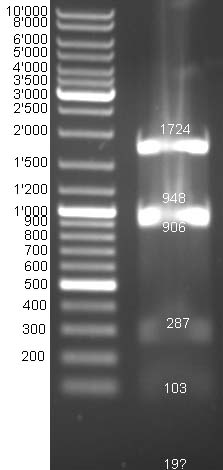
- Gel of E1 and E9 digestion: no fragments were visible
- Gel of BBa_T9002 digestion: Expected bands cannot be differentiated because fragment sizes are too similar ~1950 bp and ~2150 bp.
- Send probes of pColE9 (colE9_prot_fw_XbaI), pColE1(colE1_prot_fw_XbaI/colE1_prot_rv_SpeI/colE1_imm_fw_SpeI) and BBa_T9002 (pSB_ins_fw/pSB_ins_rv) to GATC for sequencing.
Activity Test
- Creation of Supernatant of stressed and unstressed colicin cultures.
- Plate the 4 different supernatants and the 4 different cultures on the prepared ColE1, ColE9, MG1655 and TOP10 plates.
Sender part
pBAD-Sender Cloning
- Miniprep and Glycerolstocks of pBAD18-BBa_F1610 colony 9, 10, 11
- Send miniprep of pBAD18-BBa_F161 cloning colony 9 (VIC121/VIC122 align on pBAD18)to GATC for sequencing.
Activity Test
- 24h measurement of Colicin test
[back]
Thursday 08/28/2008
Colicin Receiver
pSB1A3-Receiver-Colicin-Cloning
- Sequencing results: ColE1_prot correct. Others were not sequenced.
- New attempt of second digestion: no bands are visible.
- New cloning strategy:
- Cloning of AHL-receiver without GFP into pSB1A3 (Amplification from BBa_T9002 with T9002_XbaI_fw and T9002_SpeI_BamHI_RBS_rv primers)
- Cloning of colicin operon behind LuxPr promoter.
- In addtion:
- PCR of BBa_T9002 with new primers standardprotocol Phusion MasterMix (Finnzymes, NEB)
25.0 µl Phusion MasterMix 2.5 µl Primer fw 2.5 µl Primer rv 2.0 µl DNA Template 18.0 µl H2O ------- 50.0 µl
- PCR Purification and Analytical gel -> right fragment size
- Digestion of PCR product and vector with XbaI and SpeI
- Gelextraction: expected bands (~1061 bp LuxpR-receiver and ~2157 bp pSB1A3-backbone) could be separated and were eluted in 34 µl H2O.
- Sapping of vector to avoid self-ligation: 30 min 37 °C -> 15 min 65 °C
34.0 µl pSB1A3 DNA (XbaI/SpeI cutted) 4.0 µl SAP Buffer (Fermentas) 1.0 µl SAP enzymes (Fermentas)
- Ligation: 16 °C -> Overnight
6.0 µl Receiver DNA 2.0 µl Vector DNA 2.0 µl Buffer T4 DNA Ligase 8.0 µl H2O ------- 20.0 µl
Activity Test
- Results: No effects were obtained
- Preparation of probes for inocculation over night to measure colicin E1 activity:
- 1 x TOP 10
- 1 x TOP 10 + 2 ml ColE1 supernatant unstressed
- 1 x TOP 10 + 2 ml ColE1 supernatant stressed
- 1 x TOP 10 + 2 ml ColE9 supernatant unstressed
- 1 x TOP 10 + 2 ml ColE9 supernatant stressed
- 1 x TOP 10 + 2 ml MG1655
Sender part
pBAD-Sender Cloning
- Sequencing results: pBAD18-BBa_F1610 is the right construct.
[back]
Friday 08/29/2008
Colicin Receiver
pSB1A3-Receiver-Colicin-Cloning
- Transformation of ON-Ligation
Activity Test
Results:
- OD Measurement of colicin treated ONCs. An effect in cultures of Mytomycin C stressed cells were observed. But this could also be the toxic effect of Mytomycin C. In addition we obtained that we used a too high concentration of Mytomycin C (1 µg/ml instead of 0.1 µg/ml).
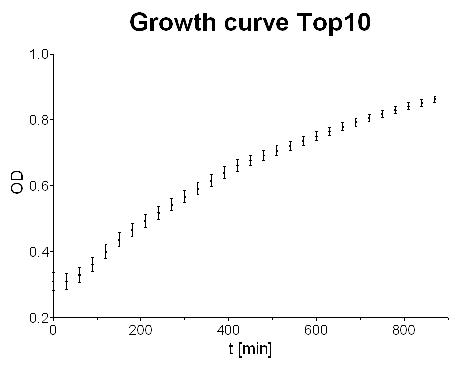
- Measured cell growth curve over night: Logistic growth could be observed but in general growth curve does not look that good.
Sender part
General
- Seminar on Synthetic Biology: Chris
- Breakfast
- Project Presentation for Frankfurter Allgemeine Zeitung
- Campus TV
- Team Meeting
[back]
 "
"