Home
People
Project Details
Protocols
Completed Systems
Biosafety
Support
|
Subprojects
Note: Click on the subproject title or picture for a detailed description of the subproject
|
|
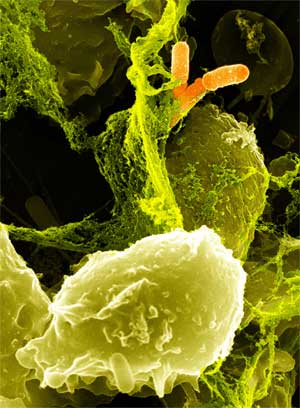
Specialized white blood cells called neutrophils defend us from illness by killing bacteria with a potent concoction of degradative enzymes and oxidizing agents, including hydrogen peroxide. However, pathogens of the human large intestine are able to cause serious illness while being sheltered from neutrophils. We engineered a strain of Escherichia coli that is able to mimic a neutrophil by producing cytotoxic amounts of hydrogen peroxide in a controlled, inducible manner. Our engineered E. coli use the transcriptional activator LuxR to detect the presence of acyl-homoserine lactones, quorum sensing signaling molecules secreted by invading pathogens. LuxR activates production of the pyruvate oxidase of Streptococcus pneumoniae, which produces large amounts of hydrogen peroxide by oxidizing pyruvate. The engineered E. coli is capable of killing certain strains of antibiotic resistant E. coli within six hours. When translated into a probiotic strain such as Nissle 1917, this system has the potential to be an effective means of combating enteric pathogens.
|
|
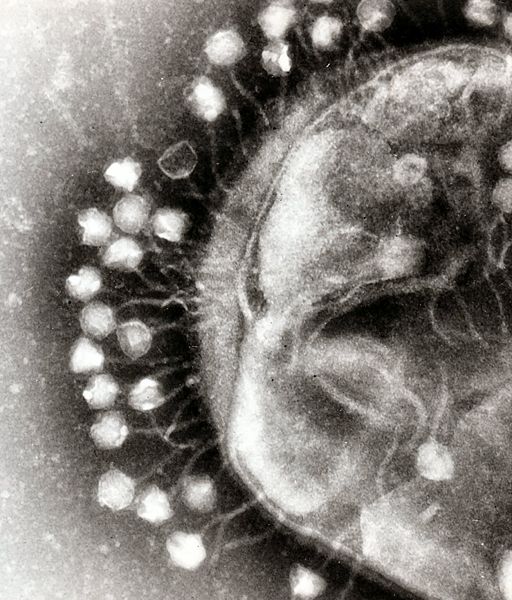
Bacterial food poisoning is a prevalent problem around the world; in the year 2006, the United States had more than 325,000 hospitalizations from food borne illnesses. The iGEM 2008 team is designing a probiotic for medical applications, and given the prevalence of foodborne illnesses, pathogen defense is high on the list of priorities. A model system for ‘manufacturing’ phage was built around Escherichia coli bacteriophage λ lysogens. A strain of E. coli has been developed which is immune to λ phage infection, but is lysogenic for λ. Furthermore, a plasmid was created to control the induction of λ using the E. coli gene rcsA. Due to the fact that the probiotic is immune to λ phage infection, the λ phage will only target pathogenic E. coli. This system can be modified to target other pathogenic species by replacing λ phage with other temperate bacteriophages and use similar methods to incorporate the phage into E. coli.
|
|
|
|
Approximately 75% of adults worldwide suffer from lactose intolerance, the inability to metabolize lactose in the small intestine. We propose to treat lactose intolerance by engineering a strain of Escherichia coli that can reside in the large intestine. The engineered strain will sense lactose and subsequently release ß-galactosidase to convert lactose into glucose and galactose, both of which can be reabsorbed by the host. To treat lactose intolerance, our engineered bacterial strain will contain two plasmids: one with constitutive expression of a mutant lactose permease and ß-galactosidase, and the second with lactose-inducible expression of the λ phage lysis cassette. The mutant lactose permease allows the cells to import lactose under all conditions. When the cells uptake enough lactose, the second plasmid will induce cell lysis through activation of the λ phage lysis cassette, resulting in cell lysis and release of ß-galactosidase into the large intestine. Data covering the construction and characterization of these plasmid constructs is discussed.
|
|
Folate, a term which encompasses the various forms of the vitamin B9, is an essential vitamin involved in everyday cell functions such as DNA replication. Unable to naturally produce folate, humans must obtain it from vegetables or folate-supplements. In regions with little or no access to these foods, folate deficiencies can cause serious birth defects. One possible solution to alleviate the effects of folate deficiency is to engineer a strain of gut microbes to produce bioavailable folate directly in the colon. We tested a total of four heterologous genes, two from the folate biosynthesis gene cluster and two from the paraaminobenzoic acid (pABA) synthesis pathway. Using standardized genetic sequences, folate biosynthesis genes extracted from the Lactoccocus lactis genome were cloned into Biobricks plasmids, transformed into Escherichia coli and overexpressed. We measured the effects of overexpression
in terms of total folate and paraaminobenzoic acid levels. PABA, an intermediate in folate synthesis, was detected using high performance liquid chromatography (HPLC). Folate detection was achieved via a microbiological assay. A measurable increase in folate production in E. coli provides proof-of-concept for both the feasibility of engineering overproduction of folate in E. coli, as well as using standardized genetic components to do so.
|
|
|
|
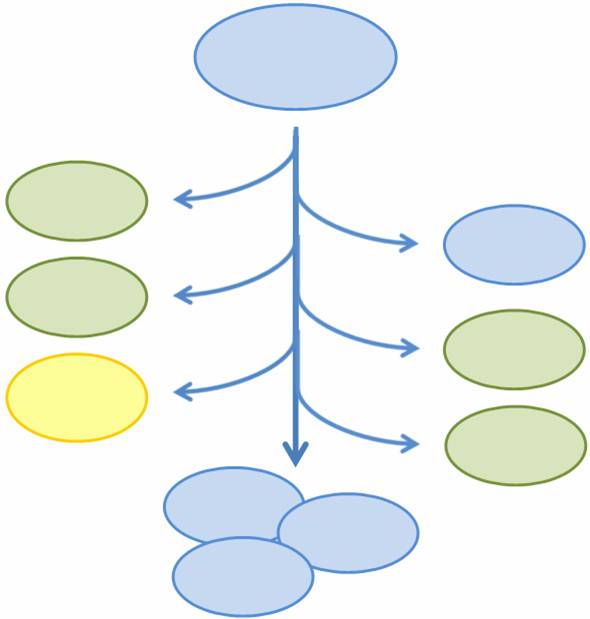
As more complicated and interconnected biological circuits are built, there is an increasing need for the simple integration of multiple functions into a single bacterial cell line. However, some of these functions may be incompatible or may kill the cell, such that each cell can only express a single function at any time or must be regenerated if the functionality is fatal. We aim to combine multiple mutually exclusive and potentially fatal functions into a single bacterial cell line that, as a population, exhibits the entire set of functions.
In particular, we want only one of the four subprojects described above to be turned on in any given cell, or else the cell may be overburdened by our constructs. At the same time, we want our bacterial population to have the capability to exhibit all four functions. In addition, three of the four subprojects result in the death of the host cell through different methods of self-induced lysis. Therefore, we need a system that is able to combine all subprojects into one coherent system and that allows for self-renewal of the population.
We propose a system in which a single bacterial strain encodes all four functions in its genome and can access each function independently through probabalistic molecular events. The engineered bacterial cells start in an undifferentiated state and randomly differentiate into one of three possible final states. To build this system, we designed two genetic devices. One relies on DNA polymerase slippage upon replication of a long stretch of short nucleotide repeats, a phenomenon termed slipped-strand mispairing (SSM). The other device uses the recombinase protein FimE to flip DNA segments. The two devices can form a system that permits the novel introduction of random multi-state differentiation into bacterial cells. Our experimental results demonstrate that short nucleotide repeats can be used as a stochastic switch and that FimE activity depends on the length of the segment being flipped.
|
  |
 "
"