Team:Caltech/Project/Phage Pathogen Defense
From 2008.igem.org
m |
|||
| (31 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
<br> | <br> | ||
__NOTOC__ | __NOTOC__ | ||
| + | |||
==Introduction== | ==Introduction== | ||
[[Image:PathogenicEcoli.jpg|thumb|left|Pathogenic ''E. coli''.]] | [[Image:PathogenicEcoli.jpg|thumb|left|Pathogenic ''E. coli''.]] | ||
| - | There are < | + | There are 10<sup>14</sup> bacterial cells that naturally reside in the human gut, an order of magnitude greater than all the cells in the body. Humans enjoy a mutualistic relationship with intestinal microbiota, wherein the microorganisms perform a host of useful functions, such as processing unused energy substrates, training the immune system, and inhibiting growth of harmful bacterial species [1]. However, not all bacteria populations within the human intestine are benign. There are many pathogenic bacteria which cause a wide variety of diseases when present in the human gut. As one example, cholera is one of the most widespread and damaging of such infectious microorganisms, with the World Health Organization reporting 132,000 cases in 2006 [2]. But cholera is just one of many examples. In the United States alone, there were more than 325,000 hospitalizations from food-borne illnesses in the year of 2006 [3]. |
| - | There are many medical treatments for food-borne illnesses, the most commonly prescribed being antibiotics. Unfortunately, antibiotics are indiscriminant and lead to rapid depletion of benign bacterial populations within the intestine. Due to this | + | There are many medical treatments for food-borne illnesses, the most commonly prescribed being antibiotics. Unfortunately, antibiotics are indiscriminant and lead to rapid depletion of benign bacterial populations within the intestine. Due to this undesireable side effect, dietary supplements have been popularized which aim to introduce beneficial bacteria back into the gut after antibiotic treatment; these substances have been termed ‘probiotics’. Natural probiotics have two main advantages: they provide useful functions for the host and they competitively inhibit growth of pathogenic bacteria. |
| - | Natural probiotics do not convey any more advantages than bacteria in a healthy human intestine. Modern synthetic biology techniques should allow us to create an engineered probiotic that goes beyond the limitations of natural probiotics. The viability of such engineered probiotics within humans has already been established [4]. As part of a collaborative effort by the Caltech iGEM team to | + | |
| + | Natural probiotics do not convey any more advantages than bacteria in a healthy human intestine. Modern synthetic biology techniques should allow us to create an engineered probiotic that goes beyond the limitations of natural probiotics. The viability of such engineered probiotics within humans has already been established [4]. As part of a collaborative effort by the Caltech iGEM team to engineer a novel probiotic with improved medical applications, this project focuses on engineering a pathogen defense system within ''E. coli''. | ||
| + | |||
| + | There are many ways for the engineered probiotic to combat pathogens in the large intestine; we chose to use bacteriophages. Bacteriophages present a novel and effective agent for eliminating pathogenic bacteria. Two important factors contribute to a bacteriophage’s effectiveness: bacteriophages are highly infectious and thus highly efficient at destroying targeted populations, and bacteriophages are specific to their hosts. The latter factor is readily illustrated by the coliphage λ, which possesses a mode of infection dependent on an ''E. coli'' specific surface protein, LamB [5]. Benign, non-''E. coli'' intestinal bacteria lack this surface receptor and therefore are immune to destruction. | ||
===System Design=== | ===System Design=== | ||
| + | |||
The engineered probiotic acts as a delivery vehicle for phage into the large intestine. There are four important design goals for such as system: (1) the system releases phage into the large intestine, (2) the production of phage is regulated with a high dynamic range between a distinct on and off state, (3) the system can target a wide range of pathogenic bacteria, and (4) the system integrates well within the existing iGEM probiotic. | The engineered probiotic acts as a delivery vehicle for phage into the large intestine. There are four important design goals for such as system: (1) the system releases phage into the large intestine, (2) the production of phage is regulated with a high dynamic range between a distinct on and off state, (3) the system can target a wide range of pathogenic bacteria, and (4) the system integrates well within the existing iGEM probiotic. | ||
| - | An obvious delivery mechanism would be the adaptation of natural lysogens, a phase of the phage life cycle in which the phage integrates its DNA | + | An obvious delivery mechanism would be the adaptation of natural lysogens, a phase of the phage life cycle in which the phage integrates its DNA into the genome of the host organism. Lysogens lie dormant until disturbed by some external stimuli, but can be stimulated to enter the lytic cycle and release phage. There is one major limitation to this method: phage released will be specific to the lysogenic hosts, ''E. coli''. We can avoid this limitation by incorporating as a lysogen a phage that is not normally infectious to ''E. coli'', such as the P22 phage from ''Salmonella''. This phage could potentially target pathogenic bacteria besides ''E. coli''. |
| + | To demonstrate the feasibility of this approach, a simple model system was developed using λ phage and JW3996-1, a strain of ''E. coli'' immune to λ infection due to a ''lamB'' deletion [5]. In this model, the host is immune to infection unless LamB is expressed from a plasmid. After infection and selection for lysogens, the ''lamB'' plasmid is counterselected against to regain immunity. Extending this model to a more realistic situation, we envision using a phage that targets a pathogenic bacterial species, constitutively expressing the phage receptor within ''E. coli'' to create lysogens, and inducing the lysogens to infect non-''E. coli'' pathogenic bacteria. | ||
| - | + | We require one other plasmid which controls the induction of the lysogens to release phage into the environment. Control of this aspect of the system is vital for integration with the overall iGEM project. The induction of λ phage lysogens has been explored since 1950 [6], and serves as a model system for the study of other temperate phages. The most famous induction mechanism is ultraviolet radiation (UV). In UV induction of λ lysogens, UV induced DNA damage activates the cellular SOS mechanism, which activates the expression of RecA, a protein involved in SOS response. RecA, in turn, cleaves the λ repressor and leads to phage activation. In this study, activation of the ''recA'' pathway was not readily feasible, so an alternative effector was used to elicit induction. A secondary plasmid expressing the ''E. coli'' regulatory gene, ''rcsA'', is used as the trigger for λ phage induction. ''rcsA'' is one of several regulatory genes which provide a ''recA'' independent pathway for λ induction [7]. Overexpression of RcsA leads to λ induction, and, furthermore, RcsA appears to directly compete with the λ repressor cI [6]. In this project, ''rcsA'' is used to trigger phage production. The effectiveness and dynamic range of ''rcsA'' induction is explored. | |
===Constructs=== | ===Constructs=== | ||
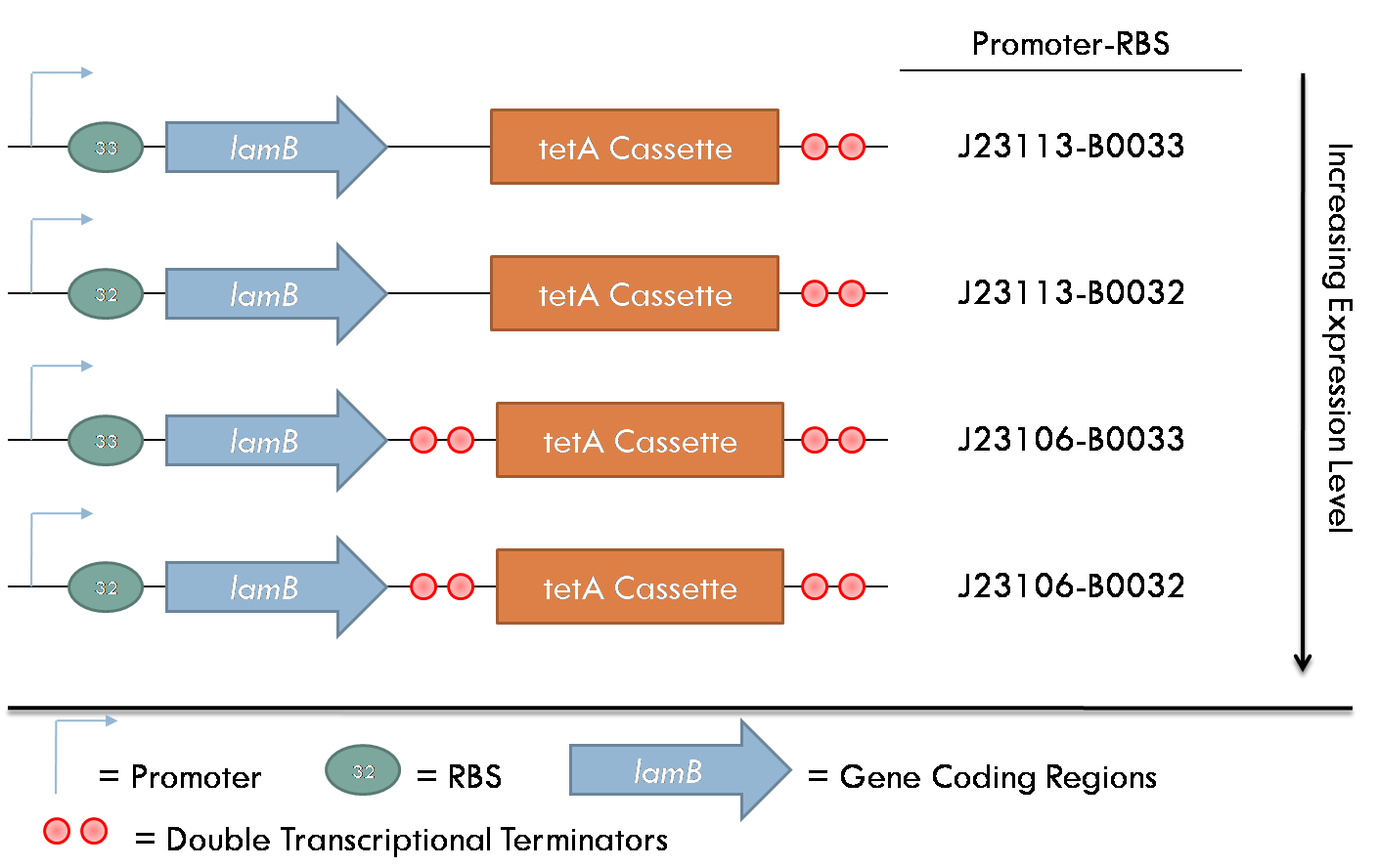
| - | ===Results=== | + | [[Image:lambteta.png|thumb|right|300px|Figure 1 - Constructs involved in expression of the ''lamB'' receptor and the TetA cassette ]] |
| + | All constructs were made with standard assembly methods. ''lamB'' and ''rcsA'' were cloned out of DH10B genomic DNA. Two families of plasmids were constructed for this project. The first consists of plasmids that express the ''lamB'' receptor and the tetracycline resistance cassette. The level of ''lamB'' expression required for λ phage infection is unknown, so the strength of the promoter/ribosome binding site (RBS) combination upstream of ''lamB'' was varied between the four constructs of this family. | ||
| + | {{clear}} | ||
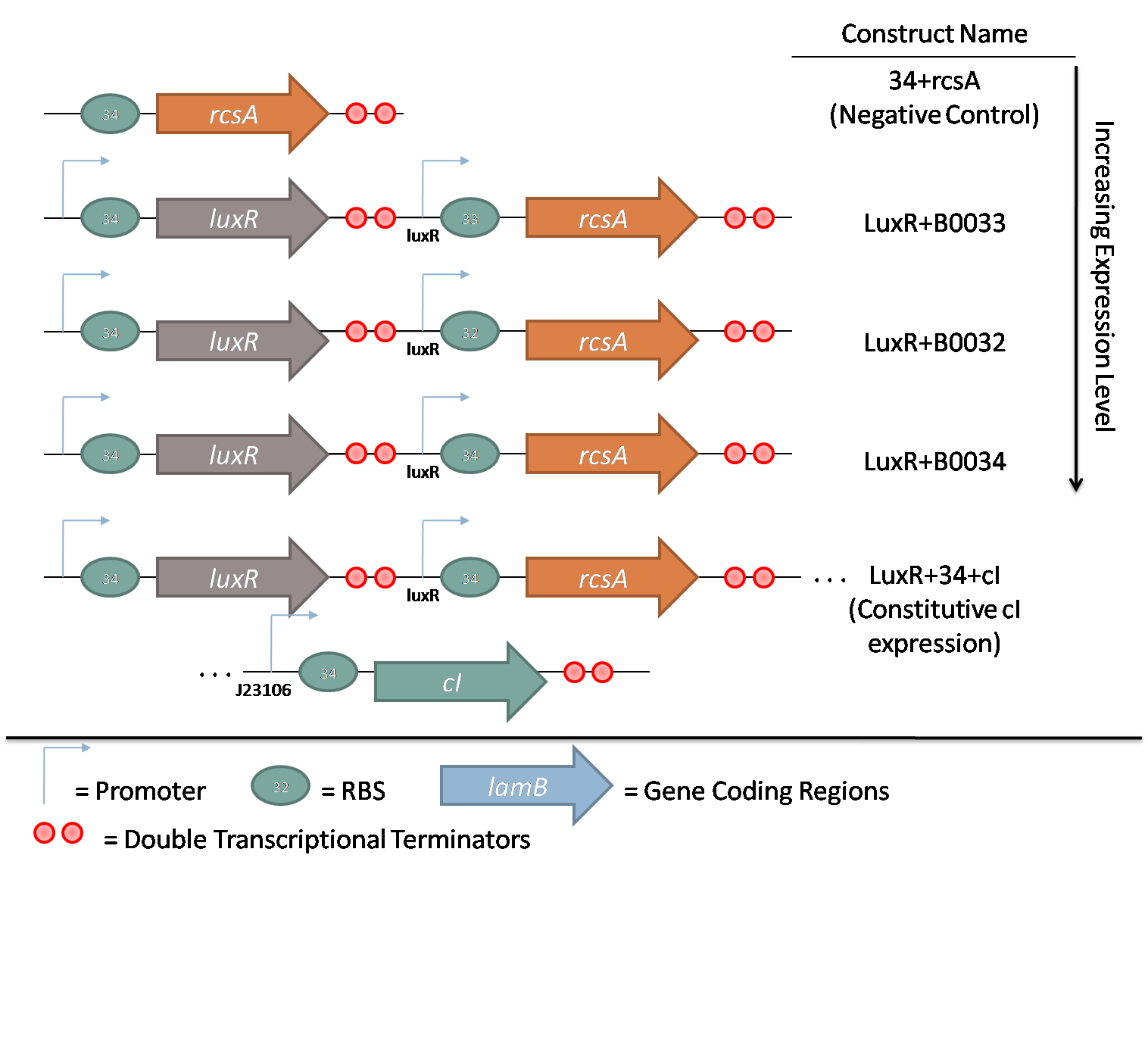
| + | [[Image:rcsa.png|thumb|left|300px|Figure 2 - Constructs for characterization of rcsA induction ]] | ||
| + | |||
| + | The second family consists of constructs used to characterize ''rcsA'' induction of λ phage. ''rcsA'' was placed downstream of an acyl-homoserine lactone (AHL) inducible promoter, LuxR. This promoter was extensively characterized by Doug for the [[Team:Caltech/Project/Population_Variation|<font style="color:#BB4400">oxidative burst project</font>]]. Using flow cytometry data, the peak response for this promoter was determined to be 10 nM AHL. Plasmids with three different RBSs in front of ''rcsA'' were constructed to characterize the response of induction. Furthermore, a promoterless ''rcsA'' construct was built as a negative control, and a construct with constitutively active λ repressor ''cI'' driven by J23106 was also assembled. | ||
| + | |||
| + | {{Clear}} | ||
| + | |||
| + | ==Results== | ||
| + | |||
| + | ===''lamB''<sup>-</sup> Infection=== | ||
| + | ''lamB'' <sup>-</sup> cells were successfully infected by λ phage while constitutively expressing the ''lamB'' receptor. Furthermore, the level of infection directly correlated with the level of ''lamB'' expression , suggesting that phage infection was limited by the presence of ''lamB'' receptors. Next, lysogens were selected using chloramphenicol resistance. The presence of the ''lamB''/tetA construct within the lysogens was confirmed with colony PCR. This plasmid was successfully removed via tetracycline counterselection [10]. This procedure successfully established a lysogenic strain lacking the ''lamB'' receptor and thus immune to phage infection. | ||
| + | |||
| + | <gallery widths="375px" heights="250px" > | ||
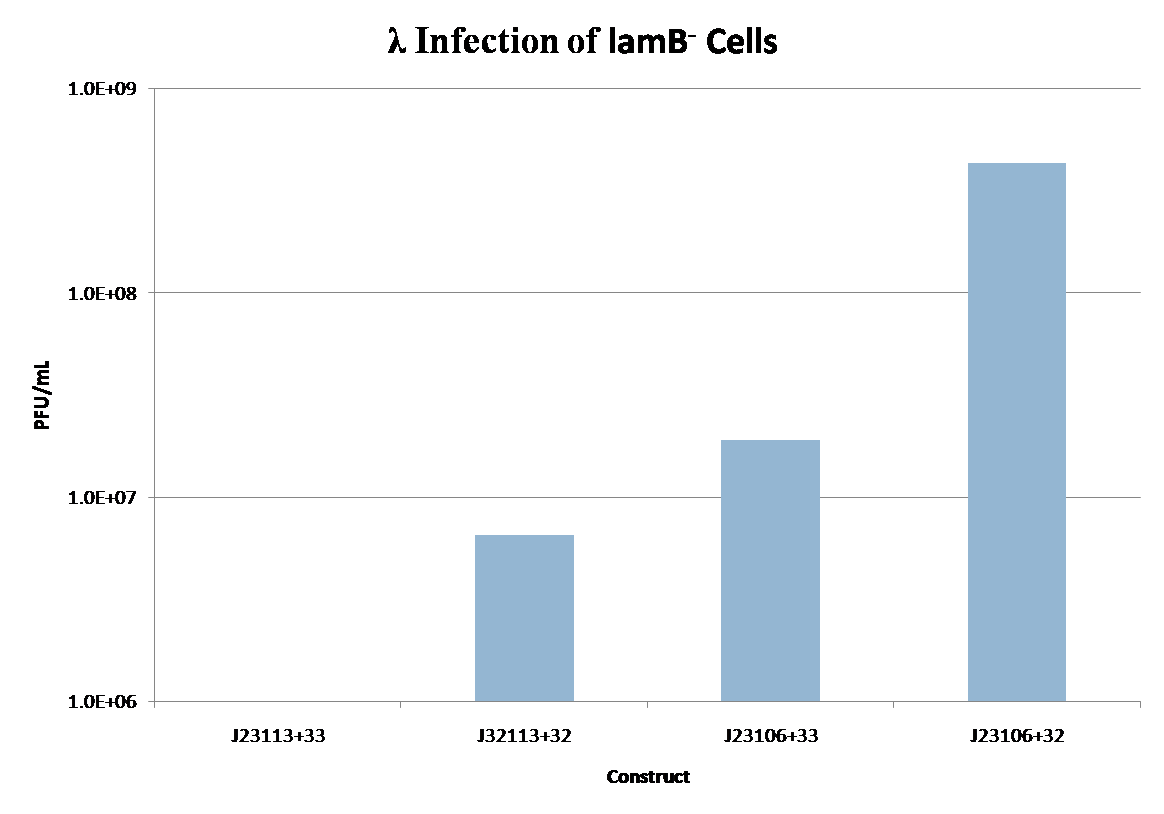
| + | Image:lamb.png|Figure 3 - λ Infection of Constitutively Active lamB Constructs. Constitutively active ''lamB'' (Table 1) was used to augment the ''lamB''- phenotype in JW3996-1 cells. Cells were titered with a known high concentration stock of phage 1x10<sup>10</sup> phage/mL. The efficiency of infection of ''lamB'' constructs can be seen through the observed phage concentration after titering. Zero plaques were observed for the J23113+33 construct. | ||
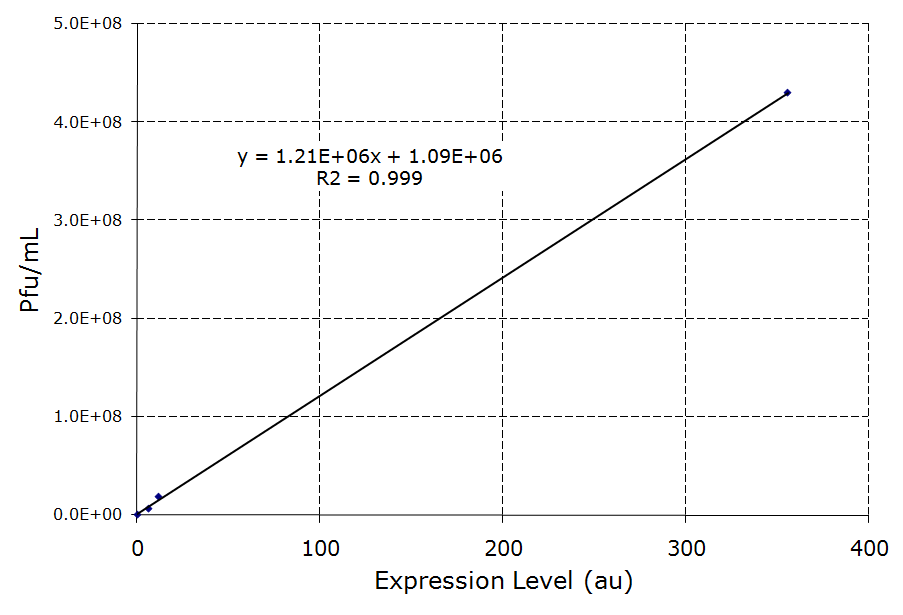
| + | Image:LamBExpression.png|Figure 4 - ''lamB'' Infection vs ''lamB'' expression level. Infection levels for the constructs in Figure 3 plotted against the expression levels recorded in the registry for the Promoter/RBSs used. | ||
| + | </gallery> | ||
| + | |||
| + | ===''rcsA'' Induction=== | ||
| + | |||
| + | |||
| + | Overexpression of ''rcsA'' led to induction of λ lysogens. ''rcsA'' was placed downstream of an AHL inducible promoter and overexpressed through induction with 10 nM AHL. Following induction, phage concentration in the supernatant was recorded through titering. This concentration was compared to the phage concentration in the supernatant without ''rcsA'' overexpression, given by a construct where ''rcsA'' is not driven by a promoter. The data show a 40 fold increase in phage concentration between the promoter-less ''rcsA'' construct and the strongest AHL induced ''rcsA'' construct (Figure 5, columns 1 and 4). The promoter-less ''rcsA'' construct should accurately represent background phage levels for the lysogen absent of any ''rcsA'' expression. | ||
| + | Further work was done to characterize the response of the AHL inducible switch under three different RBSs. The data show induction efficiency is directly correlated with strength of ''rcsA'' overexpression (Figure 5, columns 2-4). In each of the constructs, there is a 3 to 5 fold increase in phage levels after induction with 10 nM AHL. This increase does not accurately reflect the dynamic range of the AHL inducible promoter, because it does not take into account the background phage levels without the promoter. In the weaker RBSs (B0032 and B0033), the phage concentration from the uninduced supernatant is comparable to the background level from the promoter-less construct (Figure 5, columns 1-3). Thus, in these inducible promoter systems, there is high dynamic range and negligible background compared to basal phage concentrations. However, in the strongest RBS (B0034), the uninduced phage level is an order of magnitude higher than the promoter-less background level. This high basal level expression suggests that the higher expression levels due to the strong RBS is leading to leaky ''rcsA'' activation of the phage lytic cycle. | ||
| + | |||
| + | [[Image:Rcsainduction.png|600px|thumb|center|Figure 5 - ''rscA'' induction of phage production. In this figure, each construct was infected with λ phage and lysogens were selected. Lysogens were grown to OD600=0.1 and ''rcsA'' was induced with 10 nM of AHL. 1.5 hours after induction, the supernatant was used to titer wildtype stock of ''E. coli''. Plaques were counted to calculate the phage concentration inside the supernatant after induction. For more information see [[Team:Caltech/Protocols/rcsA_Lysogen_Induction|<font style="color:#BB4400">induction protocol.</font>]]]] | ||
| + | {{Clear}} | ||
| + | |||
| + | ===''cI'' reduces ''rcsA'' induction=== | ||
| + | |||
| + | The high level of background phage induction in the strongest AHL inducible ''rcsA'' switch was not optimal. If the high basal phage levels could be reduced, then the dynamic range of the inducible switch can be greatly increased. cI, the λ repressor, has been shown to directly inhibit UV induction of lysogens [13], and RcsA seems to competitively interact with cI in λ induction [7]. Both of these properties suggest that increased concentration of the λ repressor should lead to lower phage induction levels. ''cI'' was constitutively expressed downstream of the strongest AHL inducible switch. As expected, expression of cI reduces the phage release and ''rcsA'' induction in both the off-state as well as the induced state. Phage concentration was reduced by more than 1.5x106 pfu/mL in both control and induced states (Figure 5, Columns 4 and 5). In Figure 5, for the construct expressing ''cI'', there is an 8 fold increase in phage concentration between induced and uninduced states, more than double the dynamic range of the construct without ''cI'' expression. | ||
| + | |||
| + | ==Conclusion== | ||
| + | We have successfully developed a model system in which a bacteriophage can be integrated as a lysogen within an ''E. coli'' host that is not able to be infected by that phage. We successfully demonstrated this model with bacteriophage λ and the JW3996-1 strain of ''E. coli'' which is immune to λ infection. A system for inducing the release of phage was constructed around the ''E. coli'' regulatory protein RcsA. Work was done to optimize a switch using an AHL inducible promoter for ''rcsA'' induction. RcsA overexpression led to significant phage induction, producing approximately 1x10<sup>8</sup> plaque forming units (pfu)/mL. This value is 4 orders of magnitude greater than the phage induction reported by Rozanov et al. However, direct comparisons cannot be readily made between the data, because the method of overexpression of ''rcsA'' differs between the two papers, and the level at which ''rcsA'' was overexpressed in Rozanov’s study is unknown. The background phage concentration in uninduced lysogens was also much higher than reported in literature [7]. At present, there is no clear cause for this increase in basal induction levels within the λ lysogens. Phage induction via ''rcsA'' can be reduced through lambda repressor overexpression. The mechanism by which RcsA activates the lytic cycle of the dormant prophage is largely unknown; however, there is evidence that RscA acts through inactivation of the phage repressor, ''cI'' [7]. Therefore, increased production of the repressor ''cI'' led to a decrease in ''rcsA'' mediated phage induction. The addition of constitutively active ''cI'' repressor led to the construction of an ''rcsA'' phage induction system with a high dynamic range and relatively low background. | ||
| - | + | Future work should be focused on broadening the host range of the system. Thus, a key aspect needs to be adapting other temperate phages to the model system. A significant extension of the results presented here would be the incorporation and production of a non-''E. coli'' bacteriophage by ''E. coli''. A promising candidate for this is the Salmonella phage P22; P22 has been shown to be viably produced in ''E. coli'' when the prophage is expressed [14]. Incorporation of the P22 prophage should be feasible following the framework of the current model. Furthermore, co-culture experiments with the current constructs and wild-type ''E. coli'' can be used to validate the current system's effectiveness at eliminating pathogens. | |
| - | + | ==References== | |
| + | # Sears CL. 2005. "'''A dynamic partnership: Celebrating our gut flora.'''" Anaerobe, Volume 11, Issue 5, Pages 247-251. | ||
| + | # "'''Cholera Statistics 2006.'''" World Health Organization. 4 July 2008 <http://www.who.int/wer/2006/wer8131.pdf>. | ||
| + | # "'''Food Borne Illness FAQ.'''" World Health Organization. 5 July 2008 <http://www.who.int/mediacentre/factsheets/fs237/en/>. | ||
| + | # Westendorf AM, Gunzer F, Deppenmeier S, Tapadar D, Hunger JK, Schmidt MA, Buer J, and Bruder D. “'''Intestinal immunity of Escherichia coli NISSLE 1917: a safe carrier for therapeutic molecules.'''” FEMS Immunol Med Microbiol 2005 Mar 1; 43(3) 373-84. | ||
| + | # Clément JM, Hofnung M. “'''Gene sequence of the lambda receptor, an outer membrane protein of E. coli K12.'''” Cell. 1981 Dec; 27(3 Pt 2):507–514. | ||
| + | # Lwoff, A., L. Siminovitch, and N. Kjeldgaard. 1950. “'''Induction de la production de bacteriophages chez une bacterie lysogene.'''” Ann. Inst. Pasteur (Paris) 79:815-859. | ||
| + | # Rozanov, Dmitry V., Richard D'ari, and Sergey P. Sineoky. "'''RecA-Independent Pathways of Lambdoid Prophage Induction in Escherichia coli.'''" Journal of Bacteriology 180 (1998): 6306-315. | ||
| + | # “'''Biobricks Assembly'''” OpenWetware <http://openwetware.org/wiki/BioBricks_construction_tutorial> | ||
| + | # Sambrook, Joe, and David Russell. "'''Molecular Cloning.'''" New York: Cold Spring Harbor Laboratory P, 2000. | ||
| + | # Maloy, Stanley R., and William D. Nunn. "'''Selection for Loss of Tetracycline Resistance by Escherichia coli.'''" Journal of Bacteriology 45 (1985): 1110-112. | ||
| + | # Grayson, Paul. "'''Titering.'''" California Institute of Technology. <http://www.rpgroup.caltech.edu/wiki/index.php?title=titering>. | ||
| + | # Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. “'''Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection.'''” Mol Syst Biol. 2006;2:2006.0008 | ||
| + | # John Baluch, Raquel Sussman. “'''Correlation Between UV Dose Requirement for Lambda Bacteriophage Induction and Lambda Repressor Concentration'''” Journal of Virology, June 1978, P. 595-602 | ||
| + | # Botstein, David and Ira Herskowitz. "'''Properties of hybrids between Salmonella phage P22 and coliphage lambda.'''" Nature. 1974 Oct 18;251(5476):584-9. | ||
| + | }} | ||
Latest revision as of 17:00, 25 October 2008
|
People
|
Phage Pathogen Defense
IntroductionThere are 1014 bacterial cells that naturally reside in the human gut, an order of magnitude greater than all the cells in the body. Humans enjoy a mutualistic relationship with intestinal microbiota, wherein the microorganisms perform a host of useful functions, such as processing unused energy substrates, training the immune system, and inhibiting growth of harmful bacterial species [1]. However, not all bacteria populations within the human intestine are benign. There are many pathogenic bacteria which cause a wide variety of diseases when present in the human gut. As one example, cholera is one of the most widespread and damaging of such infectious microorganisms, with the World Health Organization reporting 132,000 cases in 2006 [2]. But cholera is just one of many examples. In the United States alone, there were more than 325,000 hospitalizations from food-borne illnesses in the year of 2006 [3]. There are many medical treatments for food-borne illnesses, the most commonly prescribed being antibiotics. Unfortunately, antibiotics are indiscriminant and lead to rapid depletion of benign bacterial populations within the intestine. Due to this undesireable side effect, dietary supplements have been popularized which aim to introduce beneficial bacteria back into the gut after antibiotic treatment; these substances have been termed ‘probiotics’. Natural probiotics have two main advantages: they provide useful functions for the host and they competitively inhibit growth of pathogenic bacteria. Natural probiotics do not convey any more advantages than bacteria in a healthy human intestine. Modern synthetic biology techniques should allow us to create an engineered probiotic that goes beyond the limitations of natural probiotics. The viability of such engineered probiotics within humans has already been established [4]. As part of a collaborative effort by the Caltech iGEM team to engineer a novel probiotic with improved medical applications, this project focuses on engineering a pathogen defense system within E. coli. There are many ways for the engineered probiotic to combat pathogens in the large intestine; we chose to use bacteriophages. Bacteriophages present a novel and effective agent for eliminating pathogenic bacteria. Two important factors contribute to a bacteriophage’s effectiveness: bacteriophages are highly infectious and thus highly efficient at destroying targeted populations, and bacteriophages are specific to their hosts. The latter factor is readily illustrated by the coliphage λ, which possesses a mode of infection dependent on an E. coli specific surface protein, LamB [5]. Benign, non-E. coli intestinal bacteria lack this surface receptor and therefore are immune to destruction. System DesignThe engineered probiotic acts as a delivery vehicle for phage into the large intestine. There are four important design goals for such as system: (1) the system releases phage into the large intestine, (2) the production of phage is regulated with a high dynamic range between a distinct on and off state, (3) the system can target a wide range of pathogenic bacteria, and (4) the system integrates well within the existing iGEM probiotic. An obvious delivery mechanism would be the adaptation of natural lysogens, a phase of the phage life cycle in which the phage integrates its DNA into the genome of the host organism. Lysogens lie dormant until disturbed by some external stimuli, but can be stimulated to enter the lytic cycle and release phage. There is one major limitation to this method: phage released will be specific to the lysogenic hosts, E. coli. We can avoid this limitation by incorporating as a lysogen a phage that is not normally infectious to E. coli, such as the P22 phage from Salmonella. This phage could potentially target pathogenic bacteria besides E. coli. To demonstrate the feasibility of this approach, a simple model system was developed using λ phage and JW3996-1, a strain of E. coli immune to λ infection due to a lamB deletion [5]. In this model, the host is immune to infection unless LamB is expressed from a plasmid. After infection and selection for lysogens, the lamB plasmid is counterselected against to regain immunity. Extending this model to a more realistic situation, we envision using a phage that targets a pathogenic bacterial species, constitutively expressing the phage receptor within E. coli to create lysogens, and inducing the lysogens to infect non-E. coli pathogenic bacteria. We require one other plasmid which controls the induction of the lysogens to release phage into the environment. Control of this aspect of the system is vital for integration with the overall iGEM project. The induction of λ phage lysogens has been explored since 1950 [6], and serves as a model system for the study of other temperate phages. The most famous induction mechanism is ultraviolet radiation (UV). In UV induction of λ lysogens, UV induced DNA damage activates the cellular SOS mechanism, which activates the expression of RecA, a protein involved in SOS response. RecA, in turn, cleaves the λ repressor and leads to phage activation. In this study, activation of the recA pathway was not readily feasible, so an alternative effector was used to elicit induction. A secondary plasmid expressing the E. coli regulatory gene, rcsA, is used as the trigger for λ phage induction. rcsA is one of several regulatory genes which provide a recA independent pathway for λ induction [7]. Overexpression of RcsA leads to λ induction, and, furthermore, RcsA appears to directly compete with the λ repressor cI [6]. In this project, rcsA is used to trigger phage production. The effectiveness and dynamic range of rcsA induction is explored. ConstructsAll constructs were made with standard assembly methods. lamB and rcsA were cloned out of DH10B genomic DNA. Two families of plasmids were constructed for this project. The first consists of plasmids that express the lamB receptor and the tetracycline resistance cassette. The level of lamB expression required for λ phage infection is unknown, so the strength of the promoter/ribosome binding site (RBS) combination upstream of lamB was varied between the four constructs of this family. The second family consists of constructs used to characterize rcsA induction of λ phage. rcsA was placed downstream of an acyl-homoserine lactone (AHL) inducible promoter, LuxR. This promoter was extensively characterized by Doug for the oxidative burst project. Using flow cytometry data, the peak response for this promoter was determined to be 10 nM AHL. Plasmids with three different RBSs in front of rcsA were constructed to characterize the response of induction. Furthermore, a promoterless rcsA construct was built as a negative control, and a construct with constitutively active λ repressor cI driven by J23106 was also assembled. ResultslamB- InfectionlamB - cells were successfully infected by λ phage while constitutively expressing the lamB receptor. Furthermore, the level of infection directly correlated with the level of lamB expression , suggesting that phage infection was limited by the presence of lamB receptors. Next, lysogens were selected using chloramphenicol resistance. The presence of the lamB/tetA construct within the lysogens was confirmed with colony PCR. This plasmid was successfully removed via tetracycline counterselection [10]. This procedure successfully established a lysogenic strain lacking the lamB receptor and thus immune to phage infection.
rcsA InductionOverexpression of rcsA led to induction of λ lysogens. rcsA was placed downstream of an AHL inducible promoter and overexpressed through induction with 10 nM AHL. Following induction, phage concentration in the supernatant was recorded through titering. This concentration was compared to the phage concentration in the supernatant without rcsA overexpression, given by a construct where rcsA is not driven by a promoter. The data show a 40 fold increase in phage concentration between the promoter-less rcsA construct and the strongest AHL induced rcsA construct (Figure 5, columns 1 and 4). The promoter-less rcsA construct should accurately represent background phage levels for the lysogen absent of any rcsA expression. Further work was done to characterize the response of the AHL inducible switch under three different RBSs. The data show induction efficiency is directly correlated with strength of rcsA overexpression (Figure 5, columns 2-4). In each of the constructs, there is a 3 to 5 fold increase in phage levels after induction with 10 nM AHL. This increase does not accurately reflect the dynamic range of the AHL inducible promoter, because it does not take into account the background phage levels without the promoter. In the weaker RBSs (B0032 and B0033), the phage concentration from the uninduced supernatant is comparable to the background level from the promoter-less construct (Figure 5, columns 1-3). Thus, in these inducible promoter systems, there is high dynamic range and negligible background compared to basal phage concentrations. However, in the strongest RBS (B0034), the uninduced phage level is an order of magnitude higher than the promoter-less background level. This high basal level expression suggests that the higher expression levels due to the strong RBS is leading to leaky rcsA activation of the phage lytic cycle. 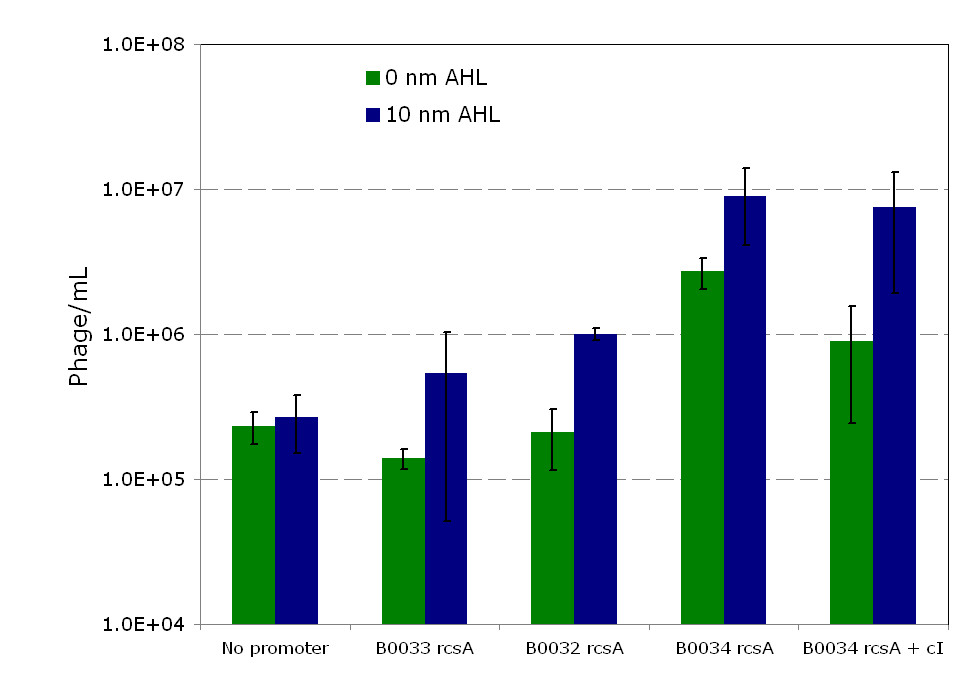 Figure 5 - rscA induction of phage production. In this figure, each construct was infected with λ phage and lysogens were selected. Lysogens were grown to OD600=0.1 and rcsA was induced with 10 nM of AHL. 1.5 hours after induction, the supernatant was used to titer wildtype stock of E. coli. Plaques were counted to calculate the phage concentration inside the supernatant after induction. For more information see induction protocol. cI reduces rcsA inductionThe high level of background phage induction in the strongest AHL inducible rcsA switch was not optimal. If the high basal phage levels could be reduced, then the dynamic range of the inducible switch can be greatly increased. cI, the λ repressor, has been shown to directly inhibit UV induction of lysogens [13], and RcsA seems to competitively interact with cI in λ induction [7]. Both of these properties suggest that increased concentration of the λ repressor should lead to lower phage induction levels. cI was constitutively expressed downstream of the strongest AHL inducible switch. As expected, expression of cI reduces the phage release and rcsA induction in both the off-state as well as the induced state. Phage concentration was reduced by more than 1.5x106 pfu/mL in both control and induced states (Figure 5, Columns 4 and 5). In Figure 5, for the construct expressing cI, there is an 8 fold increase in phage concentration between induced and uninduced states, more than double the dynamic range of the construct without cI expression. ConclusionWe have successfully developed a model system in which a bacteriophage can be integrated as a lysogen within an E. coli host that is not able to be infected by that phage. We successfully demonstrated this model with bacteriophage λ and the JW3996-1 strain of E. coli which is immune to λ infection. A system for inducing the release of phage was constructed around the E. coli regulatory protein RcsA. Work was done to optimize a switch using an AHL inducible promoter for rcsA induction. RcsA overexpression led to significant phage induction, producing approximately 1x108 plaque forming units (pfu)/mL. This value is 4 orders of magnitude greater than the phage induction reported by Rozanov et al. However, direct comparisons cannot be readily made between the data, because the method of overexpression of rcsA differs between the two papers, and the level at which rcsA was overexpressed in Rozanov’s study is unknown. The background phage concentration in uninduced lysogens was also much higher than reported in literature [7]. At present, there is no clear cause for this increase in basal induction levels within the λ lysogens. Phage induction via rcsA can be reduced through lambda repressor overexpression. The mechanism by which RcsA activates the lytic cycle of the dormant prophage is largely unknown; however, there is evidence that RscA acts through inactivation of the phage repressor, cI [7]. Therefore, increased production of the repressor cI led to a decrease in rcsA mediated phage induction. The addition of constitutively active cI repressor led to the construction of an rcsA phage induction system with a high dynamic range and relatively low background. Future work should be focused on broadening the host range of the system. Thus, a key aspect needs to be adapting other temperate phages to the model system. A significant extension of the results presented here would be the incorporation and production of a non-E. coli bacteriophage by E. coli. A promising candidate for this is the Salmonella phage P22; P22 has been shown to be viably produced in E. coli when the prophage is expressed [14]. Incorporation of the P22 prophage should be feasible following the framework of the current model. Furthermore, co-culture experiments with the current constructs and wild-type E. coli can be used to validate the current system's effectiveness at eliminating pathogens. References
|
 "
"