Team:Cambridge/Bacillus
From 2008.igem.org
Tasslehoff (Talk | contribs) |
|||
| (12 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{Cambridge/Notitle}} | |
| - | <div | + | <div class=bodytable> |
| - | { | + | {{Cambridge08}} |
| - | + | {{Cambridge08bacillus}} | |
| - | + | ||
| - | {{ | + | |
| - | + | ||
| - | + | ||
__NOTOC__ | __NOTOC__ | ||
| + | __NOEDITSECTION__ | ||
| + | <html> | ||
| + | <table style="background:#444444; padding:15px;"> | ||
| + | <table align=left width= border="0" style="background:#444444; padding:5px;"> | ||
| + | <tr> | ||
| + | <td style="width:100%; height: 400; padding-left: 15px;"> | ||
| + | <b class="b1f"></b><b class="b2f"></b><b class="b3f"></b><b class="b4f"></b> | ||
| + | <div class="contentf"> | ||
| + | <div style="height: 400; background: white; line-height:170% padding: 5px;"> | ||
| + | <div style="color: white; font: 2px;">x</div> | ||
| + | </html> | ||
| + | '''To build more complex cellular systems, new tools and techniques are required. We are generating standardized parts, tools, and techniques for the gram-positive chassis ''B. subtillis''. Easy to handle and transform, this bacterium offers many adantages to ''E. coli', including the ability to secrete proteins and integrate DNA into the chromosome. We have designed, built, and submitted gram-positive RBSes, promoters, and shuttle vectors.As a part of this work we have confirmed single copy chromosomal insertion, demonstrated InFusion assembly, and characterized an improved GFP variant. You can find our [[IGEM:Cambridge/2008/Turing Pattern Formation/Experiments/Bacillus subtilis transfomation| ''Bacillus'' protocols here]].''' | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | ==Improved GFP== | |
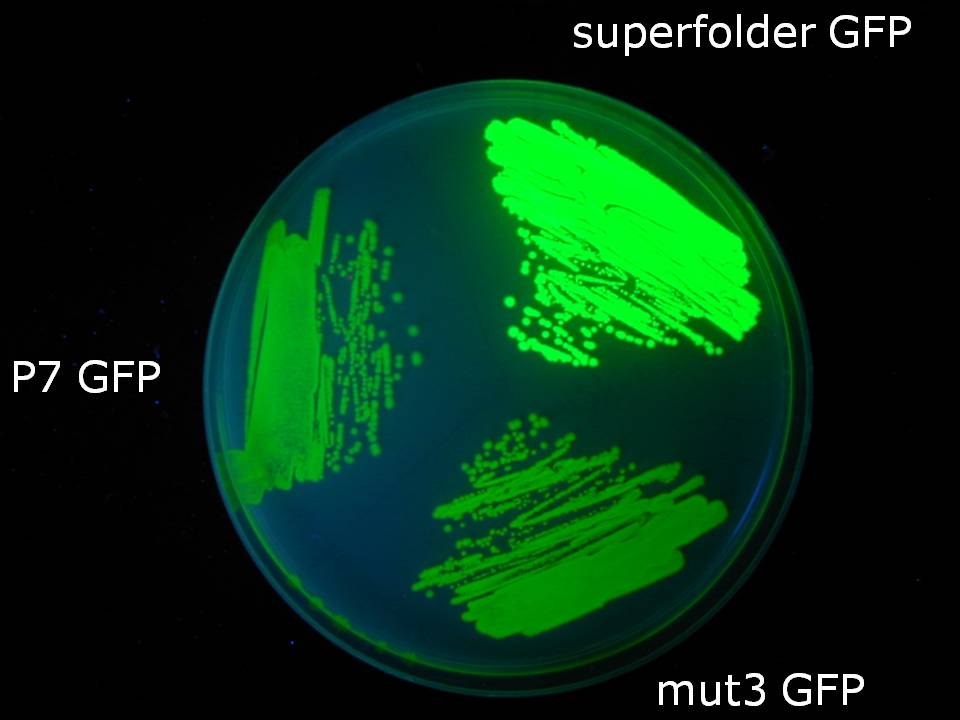
| - | = | + | [[Image:plate picture.jpg| thumb | 300px | center | Superfolder GFP along with other variants under UV light]] |
| - | + | We created a codon-optimized and biobricked version of the 'Superfolder' GFP engineered by Pédelacq et al (2006). This GFP variant folds faster and glows brighter than other GFP variants. [[Team:Cambridge/Improved_GFP|Read more...]] | |
| - | =Creating new Biobrick-compatible ''B.subtilis'' vectors= | + | ==Creating new Biobrick-compatible ''B.subtilis'' vectors== |
| - | + | [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2008&group=Cambridge Vectors submitted:] | |
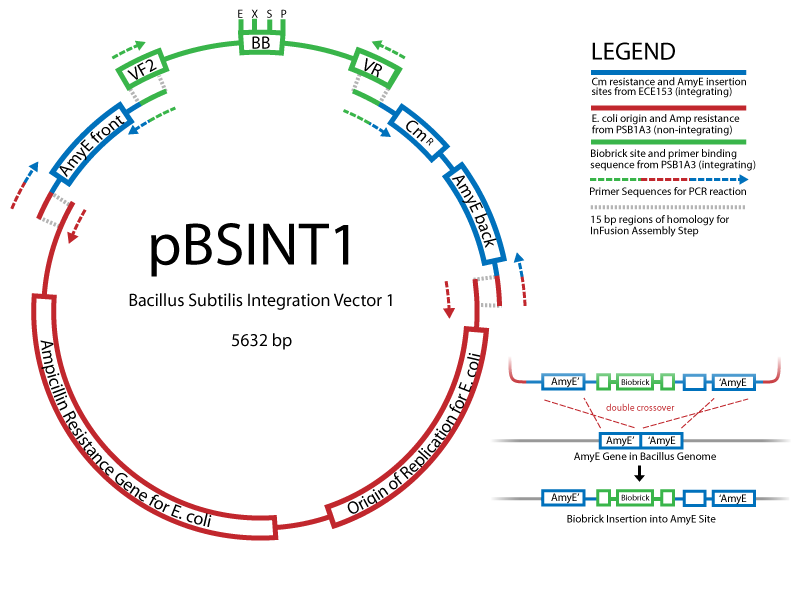
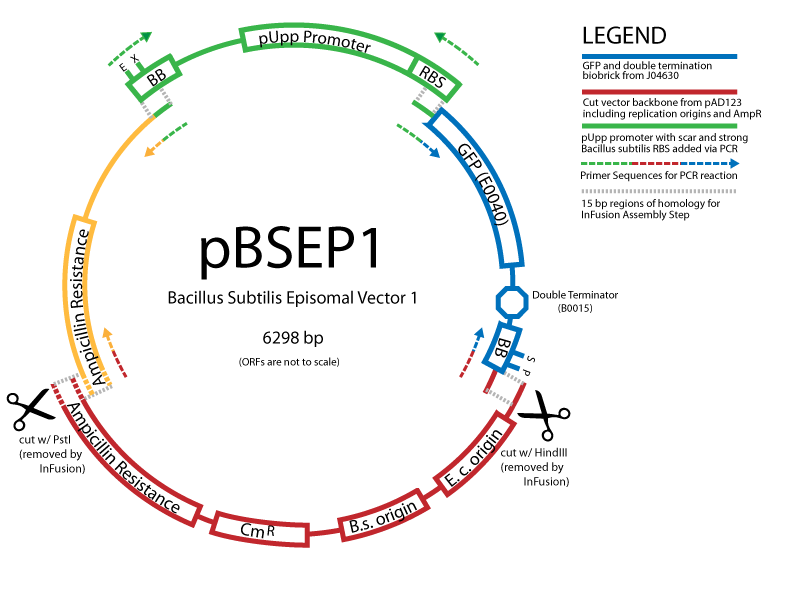
| - | We are using the In-Fusion™ PCR method from ClonTech to create new Biobrick-compatible integrative and episomal vectors. These vectors will allow us to insert Biobricks into the ''Bacillus subtilis'' genome at the AmyE locus and as a 3-5 copy plasmid that will replicate autonomously in ''Bacillus''. | + | We are using the [http://bioinfo.clontech.com/infusion/ In-Fusion™ PCR method from ClonTech] to create new Biobrick-compatible integrative and episomal vectors. These vectors will allow us to insert Biobricks into the ''Bacillus subtilis'' genome at the AmyE locus and as a 3-5 copy plasmid that will replicate autonomously in ''Bacillus''. |
{| class="wikitable" border="0" align="center" | {| class="wikitable" border="0" align="center" | ||
| Line 34: | Line 39: | ||
'''ECE166''' : High-level constitutive expression of a Green Fluorescent Protein (with the promoter Pupp) | '''ECE166''' : High-level constitutive expression of a Green Fluorescent Protein (with the promoter Pupp) | ||
| + | [[Team:Cambridge/Bacillus_subtilis_transformation|Read more...]] | ||
| + | |||
[[Image: phototrans166.jpg | thumb | 300px | center | Observation with microscope (x20) : B.S. transformed with ECE166 ]] | [[Image: phototrans166.jpg | thumb | 300px | center | Observation with microscope (x20) : B.S. transformed with ECE166 ]] | ||
| Line 43: | Line 50: | ||
[[Image: 153Amylase.jpg | thumb | 200px | left | IA751 transformed with ECE153 : SUCCESFUL TRANSFORMATION, no zone of clearing ]] | [[Image: 153Amylase.jpg | thumb | 200px | left | IA751 transformed with ECE153 : SUCCESFUL TRANSFORMATION, no zone of clearing ]] | ||
[[Image: 112Amylase.jpg | thumb | 200px | center | IA751 transformed with ECE112 : SUCCESFUL TRANSFORMATION for 4 colonies out of 5, no zone of clearing ]] | [[Image: 112Amylase.jpg | thumb | 200px | center | IA751 transformed with ECE112 : SUCCESFUL TRANSFORMATION for 4 colonies out of 5, no zone of clearing ]] | ||
| + | [[Team:Cambridge/Bacillus_subtilis_transformation|Read more...]] | ||
| - | + | =='''B. subtilis Promoters Designed'''== | |
| - | = | + | [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2008&group=Cambridge Biobricks submitted:] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
* Pupp - a strong constitutive ''Bacillus cereus'' promoter present on ''B. subtilis'' vector ECE166 | * Pupp - a strong constitutive ''Bacillus cereus'' promoter present on ''B. subtilis'' vector ECE166 | ||
| + | ** [[Team:Cambridge/Bacillus_subtilis_transformation| Successfully expressed in an episomal vector...]] | ||
* Ppac - constitutive, present on ''B. subtilis'' vector ECE151 | * Ppac - constitutive, present on ''B. subtilis'' vector ECE151 | ||
* Pspac - IPTG inducible, present on ''B. subtilis'' vector ECE151 | * Pspac - IPTG inducible, present on ''B. subtilis'' vector ECE151 | ||
* Pxyl - xylose inducible, present on ''B. subtilis'' vector ECE153 | * Pxyl - xylose inducible, present on ''B. subtilis'' vector ECE153 | ||
| + | [[Team:Cambridge/Signalling/Constructs| Read more about the constructs made...]] | ||
| + | [[Team:Cambridge/Signalling/Primers| Read more about the primers used...]] | ||
| - | Information about the ''B. subtilis'' vectors can be found [ | + | Information about the ''B. subtilis'' vectors can be found [[Team:Cambridge/Signalling/Vectors|here]]. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <html> | ||
| + | <div style="color: white; font: 2px;">x</div> | ||
| + | </div></div> | ||
| + | <b class="b4f"></b><b class="b3f"></b><b class="b2f"></b><b class="b1f"></b> | ||
| + | </td></tr></table></table> | ||
| + | </html> | ||
__NOEDITSECTION__ | __NOEDITSECTION__ | ||
__NOTOC__ | __NOTOC__ | ||
Latest revision as of 04:25, 30 October 2008
|
x
To build more complex cellular systems, new tools and techniques are required. We are generating standardized parts, tools, and techniques for the gram-positive chassis B. subtillis. Easy to handle and transform, this bacterium offers many adantages to E. coli', including the ability to secrete proteins and integrate DNA into the chromosome. We have designed, built, and submitted gram-positive RBSes, promoters, and shuttle vectors.As a part of this work we have confirmed single copy chromosomal insertion, demonstrated InFusion assembly, and characterized an improved GFP variant. You can find our Bacillus protocols here.
Improved GFPWe created a codon-optimized and biobricked version of the 'Superfolder' GFP engineered by Pédelacq et al (2006). This GFP variant folds faster and glows brighter than other GFP variants. Read more... Creating new Biobrick-compatible B.subtilis vectors[http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2008&group=Cambridge Vectors submitted:] We are using the [http://bioinfo.clontech.com/infusion/ In-Fusion™ PCR method from ClonTech] to create new Biobrick-compatible integrative and episomal vectors. These vectors will allow us to insert Biobricks into the Bacillus subtilis genome at the AmyE locus and as a 3-5 copy plasmid that will replicate autonomously in Bacillus.
Successful transformation of BacillusTransformation with episomal vectorsECE166 : High-level constitutive expression of a Green Fluorescent Protein (with the promoter Pupp) Read more...
Transformation with integration vectorsECE153 and ECE112 : Transformation in IA751 : Amylase test
B. subtilis Promoters Designed[http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2008&group=Cambridge Biobricks submitted:]
Read more about the constructs made... Read more about the primers used... Information about the B. subtilis vectors can be found here.
x
|
 "
"