Team:Cambridge/Signalling/Lab Work
From 2008.igem.org
(→August, 23rd) |
(→Plate biobricks from MIT) |
||
| Line 302: | Line 302: | ||
*Result : nothing, just the primers! Problem with our PCR | *Result : nothing, just the primers! Problem with our PCR | ||
| + | |||
==Plate biobricks from MIT== | ==Plate biobricks from MIT== | ||
| Line 312: | Line 313: | ||
*C0061 | *C0061 | ||
*R0063 | *R0063 | ||
| + | |||
| + | =September, 2nd= | ||
| + | |||
| + | ==Check transformation in E.coli of our ligated products== | ||
| + | |||
| + | - Single colony PCR with VF and VR : add 10μL of SDW, 5μL of MM, 1μL of VF, 1μL of VR and 3μL of cells | ||
| + | |||
| + | {|class="wikitable" style="text-align:center" border="1" | ||
| + | |- | ||
| + | ! inserted products !! number of colonies to check (chemical transf.) !! number of colonies to check (electrop. transf.) | ||
| + | |- | ||
| + | | RBS S || 3 || 2 | ||
| + | |- | ||
| + | | RBS W || 3 || 2 | ||
| + | |- | ||
| + | | Pspac || 2 || 2 | ||
| + | |- | ||
| + | | Pupp || 2 || 2 | ||
| + | |- | ||
| + | | agrD || 3 || 2 | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | - Gel (3% of agarose) | ||
| + | |||
| + | [[Image:gel1.gif|200px|center]] | ||
| + | |||
| + | * Result | ||
| + | |||
| + | {|class="wikitable" style="text-align:center" border="1" | ||
| + | |- | ||
| + | ! gel !! lane !! inserted product (name of the colony) !! observation !! conclusion | ||
| + | |- | ||
| + | | top || 3 || RBS S (1) || one band (260bp) || size of the insert, RBS too small to see on a gel | ||
| + | |- | ||
| + | | top || 4 || RBS S (2) || one band (260bp) || size of the insert, RBS too small to see on a gel | ||
| + | |- | ||
| + | | top || 5 || RBS S (4) || nothing || ? | ||
| + | |- | ||
| + | | top || 6 || RBS S (5) || nothing || ? | ||
| + | |- | ||
| + | | top || 7 || RBS W (1) || 2 band (260bp + 350bp) || size of the insert, RBS too small to see on a gel + something else? | ||
| + | |- | ||
| + | | top || 8 || RBS W (2) || one band (350bp) || too big | ||
| + | |- | ||
| + | | top || 9 || RBS W (4) || one band (260bp) || size of the insert, RBS too small to see on a gel | ||
| + | |- | ||
| + | | top || 10 || RBS W (5) || one band (260bp) || size of the insert, RBS too small to see on a gel | ||
| + | |- | ||
| + | | top || 11 || Pspac (1) || 2 band (260bp + 350bp) || maybe problem with products loaded on gel (exactly the same bands than for RBS W) | ||
| + | |- | ||
| + | | top || 12 || Pspac (2) || one band (450bp) || size of Pupp? | ||
| + | |- | ||
| + | | top || 13 || Pspac (3) || one band (450bp) || size of Pupp? | ||
| + | |- | ||
| + | | bottom || 3 || Pspac (4) || nothing || ? | ||
| + | |- | ||
| + | | bottom || 4 || Pupp (1) || one band (400bp) || size of Pspac? | ||
| + | |- | ||
| + | | bottom || 5 || Pupp (2) || one band (400bp) || size of Pspac? | ||
| + | |- | ||
| + | | bottom || 6 || Pupp (3) || one band (slightly lower than 400bp) || size of Pspac? | ||
| + | |- | ||
| + | | bottom || 7 || Pupp (4) || one band (400bp) || size of Pspac? | ||
| + | |- | ||
| + | | bottom || 8 || agrD (1) || 2 bands (380 and 450bp) || big band is ok | ||
| + | |- | ||
| + | | bottom || 9 || agrD (2) || 2 bands (380 and 450bp) || big band is ok | ||
| + | |- | ||
| + | | bottom || 10 || agrD (3) || no bands || ? | ||
| + | |- | ||
| + | | bottom || 11 ||agrD (4) || strong band (450bp) || ok | ||
| + | |- | ||
| + | | bottom || 12 || P2 || one band (350bp) || ok | ||
| + | |- | ||
| + | |} | ||
| + | |||
<!-- ## Do not edit below this line unless you know what you are doing. ## --> | <!-- ## Do not edit below this line unless you know what you are doing. ## --> | ||
Revision as of 17:50, 28 October 2008
August, 16th
Agr A and C ligation to pSB4C5
Ligation was performed using Fermentas Rapid Ligation kit according to their protocol.
After ligation, samples were visualized on a gel, but little product of the correct size was seen.
For agrA we should have gotten a band of size 3700 (3000plasmid + 700 insert)
For agrC we should have gotten a band of size 4300 (3000plasmid + 1300insert)
The bands of appropriate sizes were extracted for transformation.
August, 18th
Check promoters
On Friday, PCR out promoters, we want to check them.
- Run a gel
- Results : good size of bands!!!
Transformation of AgrA and AgrC
AgrA and C gell-extracted ligation was transformed into Top 10 cells using standard protocol. Puc9 was used as a positive control.
August, 19th
Results of AgrA and C transformation
Transformation has failed. No colonies were visible for the plates of Agr A or C. The Puc9 positive control grew. We believe that the problem was in the gel-extraction between ligation and transformation. Most of our plasmid was probably lost in this step. Next time we will directly use the results from the ligation reaction to transform. Although many bands were seen on the previous gel of the ligation reaction, we will check our transformation growth by single colony PCR to confirm transformation of the plasmid with our correct insert.
Lux parts
To make the Lux Receiver, we need 4 different parts ;
- R0040, TetR repressible promoter
- SO168, luxR + double terminator
- R0062, promoter activated by luxR
- JO4630 (GFP + double terminator)
All these parts have been transformed into E.coli. We want to test them. R004, R0062 and JO4630 have already been tested, it should be fine. We received from the MIT R0040, R0062 and S068 already transformed into E.coli, so we want to check these stocks (which are certainly fine) and use them. For JO4630, we want to double check our transformation.
- Plate on antibiotic plates and do LB stocks of single colony from the MIT stock (R0040, R0062 and S0168).
- Put on Kan plates 4 different colonies from J04630 (transformation Amp plate) and also incubate these colonies into LB
August, 20th
Check Lux components
- Single colony PCR for :
- R0040 (MIT stuff)
- R0062 (MIT stuff)
- 4 different colonies of S0168 (from a transformation plate from 12/08)
- 4 different colonies of J04630 (from a transformation plate from 12/08)
- Protocol : add 1μL of cells (diluted in water), 10μL of Master Mis, 7μL of SDW, 1μL of VF primer and 1μL of VR primer
- Gel PCR products
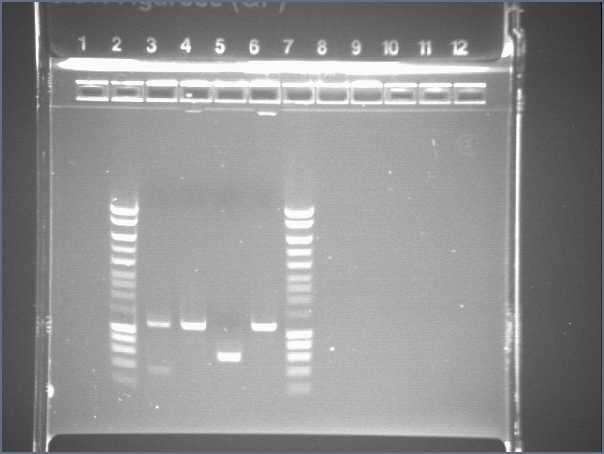
Gel 1
- Lane2 : Hyperladder1
- Lane3 : JO4630, colony 1
- Lane4 : JO4630, colony 2
- Lane5 : JO4630, colony 3
- Lane6 : JO4630, colony 4
- Lane7 : HyperladderI
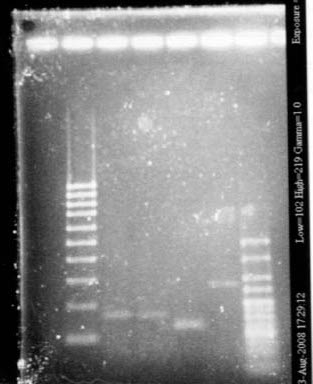
Gel 2
- Lane2 : HyperladderI
- Lane3 : ECE190 double digest
- Lane4 : S0168, colony 1
- Lane5 : S0168, colony 2
- Lane6 : S0168, colony 3
- Lane7 : S0168, colony 4
- Lane8 : HyperladderI
- Lane9 : R0040
- Lane10 : R0062
- Lane11 : Ladder 100bp
- Results
- R0040 and R0062 : one big band of about 300b (expected size 293), OK!
- S0168 : one band of about 400b for the 4 different colonies (expected size 1234!), bad! This plate does not contain S0168
- J04630 (colonies 2 and 4) : one band of about 1100b (expected size 1173), OK!
- J04630 (colony 1) : one good band plus another band...
- J04630 (colony 3) : one band of about 600b, bad!
Ligation
- Materials :
- AgrA
- AgrB
- AgrC
- AgrD
- Pupp
- Pspac
- Ppac
- Pxyl
- RBS S
- RBS W
- psB4C5
- Double digest of PCR products
- Run vector, AgrA and AgrD on a gel
- DNA clean and concentrator for AgrA, B,C and D, promoters
- Microclean for both RBS
- Nanodrop
| 260/280 | ng/μL | |
|---|---|---|
| AgrA | 1.66 | 16.4 |
| AgrB | 1.91 | 23.5 |
| AgrC | 1.99 | 35.9 |
| AgrD | 2.13 | 4.9 |
| Pxyl | 1.54 | 5.6 |
| Ppac | 1.49 | 4.6 |
| Pspc | 1.62 | 9.6 |
| Pupp | 1.88 | 8.5 |
| RBS S | 2.44 | 29 |
| RBS W | 1.44 | 10.7 |
- Extract plasmid annd Agr from gel and clean
- Ligation
August, 21st
Transformation of ligation products
- Spin chemically competent TOP10, add 100μL of CaCl2 solution
- Add 5μL of DNA (ligation products), and 1μL of PUC9
- Continue the protocol of transformation
J04630
- Plate good colonies from yesterday on Kan25 and put in LB (for plasmid stock for tomorrow)
Plasmid stocks
- Plasmid miniprep R0040 and R0062
August, 22nd
New PCR of promoters
August, 23rd
Check promoters (after PCR from bacillus vectors)
- Gel
- Lane1 : Hyperladder 5
- Lane2 : Pxyl
- Lane3 : Pspac
- lane4 : Ppac
- Lane5 : Pupp
Size is ok.
August, 27th
New transformation of ligation products
- Products : agrD, Pupp, Pspac, RBS W, RBS S (from ligation with biolabs kit)
- Competent top 10 from the freezer
- Add 5μL of ligation products, and 1.2μL of PUC9
August, 28th
Results from transformation of ligation products
- nothing on plate, even on control plate
- Reasons ?
- cells non competent anymore (try with fresh competent cells)
- not enough DNA (try with 10μL of DNA)
- very short ligation products
Transformation of ligation products (new)
- Products to ligate : Pupp, Pspac, agrD, RBS S and RBS W
- new fresh competent TOP 10
- 5μL of DNA (1.5μL of PUC9)
- 2h30 in the incubator
August, 29th
Results of transformation with our ligation products
- Everything grew! Better efficiency with electrop. than with chemical protocol
Single colony PCR to check our transformation
| Transformed products | number of picked colonies from chemical transformation | number of picked colonies from electrop. transformation (neat) | number of picked colonies from electrop. transformation (1:10) |
|---|---|---|---|
| Pupp | 2 | 2 | 0 |
| Pspac | 2 | 2 | 0 |
| RBS S | 3 | 0 | 2 |
| RBS W | 3 | 0 | 2 |
| agrD | 3 | 2 | 0 |
- Single colony PCR : 13μL of SDW+cells, 5μL of Master Mix, 1μL of VF and 1μL of VR (and plate each single colony)
- Load a gel (1.3% agarose) : 5μL of PCR products + 1μL of dye (only 1μL of 100b ladder)
In the death plasmid, the VF-VR size is about 280b.
| Transformed products | size of the product (with cutting sites) | expected size after PCR (about) |
|---|---|---|
| Pupp | 255 | 480 |
| Pspac | 125 | 350 |
| RBS S | 56 | 280 |
| RBS W | 56 | 280 |
| agrD | 200 | 430 |
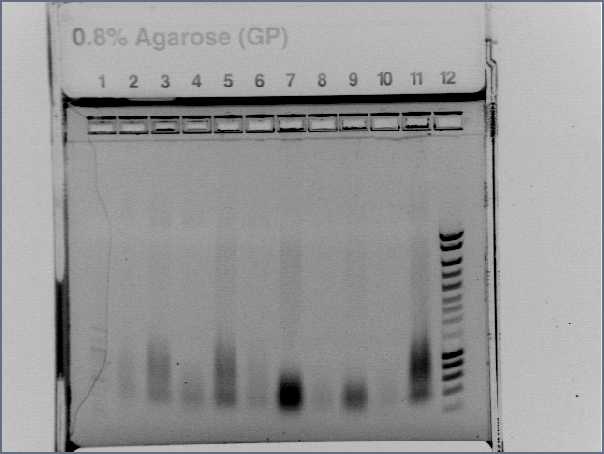
- Result : nothing, even no ladder, problem with the gel!
- run again on a e-gel
- Lane1 :ladder 100bp
- Lane2 : Pupp colony 1
- Lane3 : Pupp colony 3
- Lane4 : Pspac colony 1
- Lane5 : Pspac colony 3
- Lane6 : RBS S colony 1
- Lane7 : RBS S colony 4
- Lane8 : RBS W colony 1
- Lane9 : RBS W colony 4
- Lane10 : agrD colony 1
- Lane11 : agrD colony 4
- Lane12 : HyperladderI
- Result : nothing, just the primers! Problem with our PCR
Plate biobricks from MIT
- E0840
- B0014
- I712007
- C0012
- B0015
- C0061
- R0063
September, 2nd
Check transformation in E.coli of our ligated products
- Single colony PCR with VF and VR : add 10μL of SDW, 5μL of MM, 1μL of VF, 1μL of VR and 3μL of cells
| inserted products | number of colonies to check (chemical transf.) | number of colonies to check (electrop. transf.) |
|---|---|---|
| RBS S | 3 | 2 |
| RBS W | 3 | 2 |
| Pspac | 2 | 2 |
| Pupp | 2 | 2 |
| agrD | 3 | 2 |
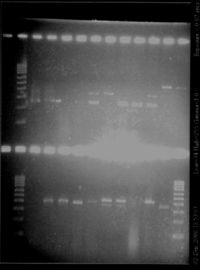
- Gel (3% of agarose)
- Result
| gel | lane | inserted product (name of the colony) | observation | conclusion |
|---|---|---|---|---|
| top | 3 | RBS S (1) | one band (260bp) | size of the insert, RBS too small to see on a gel |
| top | 4 | RBS S (2) | one band (260bp) | size of the insert, RBS too small to see on a gel |
| top | 5 | RBS S (4) | nothing | ? |
| top | 6 | RBS S (5) | nothing | ? |
| top | 7 | RBS W (1) | 2 band (260bp + 350bp) | size of the insert, RBS too small to see on a gel + something else? |
| top | 8 | RBS W (2) | one band (350bp) | too big |
| top | 9 | RBS W (4) | one band (260bp) | size of the insert, RBS too small to see on a gel |
| top | 10 | RBS W (5) | one band (260bp) | size of the insert, RBS too small to see on a gel |
| top | 11 | Pspac (1) | 2 band (260bp + 350bp) | maybe problem with products loaded on gel (exactly the same bands than for RBS W) |
| top | 12 | Pspac (2) | one band (450bp) | size of Pupp? |
| top | 13 | Pspac (3) | one band (450bp) | size of Pupp? |
| bottom | 3 | Pspac (4) | nothing | ? |
| bottom | 4 | Pupp (1) | one band (400bp) | size of Pspac? |
| bottom | 5 | Pupp (2) | one band (400bp) | size of Pspac? |
| bottom | 6 | Pupp (3) | one band (slightly lower than 400bp) | size of Pspac? |
| bottom | 7 | Pupp (4) | one band (400bp) | size of Pspac? |
| bottom | 8 | agrD (1) | 2 bands (380 and 450bp) | big band is ok |
| bottom | 9 | agrD (2) | 2 bands (380 and 450bp) | big band is ok |
| bottom | 10 | agrD (3) | no bands | ? |
| bottom | 11 | agrD (4) | strong band (450bp) | ok |
| bottom | 12 | P2 | one band (350bp) | ok |
 "
"