Team:Chiba/Experiments:LuxR mutant
From 2008.igem.org
(Difference between revisions)
(→Design) |
|||
| Line 74: | Line 74: | ||
| - | + | ||
| - | + | ||
[[Image:Mutant-LuxR Chiba.gif|frame|'''Table 1''' Seisitivity of LuxR mutants. Data modified from Collins et.al. Mol. Microbiol. 55, 712–723 (2005)]] | [[Image:Mutant-LuxR Chiba.gif|frame|'''Table 1''' Seisitivity of LuxR mutants. Data modified from Collins et.al. Mol. Microbiol. 55, 712–723 (2005)]] | ||
[[Image:AHL-activation-of-mutagenizenized LuxR molecules Chiba.gif|frame|'''Table 2''' Ahl activation of mutagenized LuxR molecules. '''Data modfied from B.Kock et al. Microbiology (2005), 151, 3589-3602''']] | [[Image:AHL-activation-of-mutagenizenized LuxR molecules Chiba.gif|frame|'''Table 2''' Ahl activation of mutagenized LuxR molecules. '''Data modfied from B.Kock et al. Microbiology (2005), 151, 3589-3602''']] | ||
| - | |||
| - | + | ||
| + | |||
'''>[[Team:Chiba/Project#Receiver|Back to the project page]]''' | '''>[[Team:Chiba/Project#Receiver|Back to the project page]]''' | ||
Revision as of 03:28, 30 October 2008
| Home | The Team | The Project | Parts Submitted to the Registry | Reference | Notebook | Acknowledgements |
|---|
LuxR mutant (Under Planning)
Design
Probably, the most straightforward approach to make variations in delay time is to create a number of LuxR mutants with different sensitivity. We thought the higher the sensitivity is, the shorter the delay time would be. The lower the sensitivity is, the slower the switch response should be.
At least, two papers on LuxR mutants (1),(2) are available. Among the many candidates, we chose the following three mutations;
- Ile45Phe: This mutation make LuxR 10x more sensitive to Lux-type AHL(1)
- Leu42Ala: Sensitivity (to AHL) becomes 1/15 of the wildtype LuxR(2)
- Leu42Ser: Sensitivity (to AHL) becomes 1/1,000 of the wildtype LuxR(2)
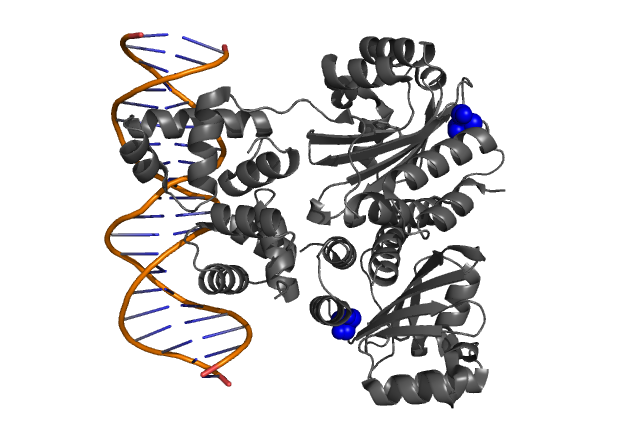
| Fig.3, Fig.4 Hypothetical positions of residues in TraR corresponding to those found to modulate acyl-HSL specificity in LuxR. The crystal structure of the LuxR homologue TraR (PDB 1L3L) has been determined(3). |
Experiment
Mutant construction:
Using sewing PCR, we introduced each of three above mutations into LuxR. Some of the parts are await for sequence verification.
Test communication:
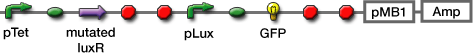
- Transformed sender (Ptet-luxI), mutant LuxR Receiver (Ptet-mLuxR-Plux-GFP) and WT LuxR Receiver into E. coli strains (JW1908)
- Separately inoculated Sender, WT Receiver (wild type luxR/BW⊿FliC) and mutated Receiver (1point mutation/BW⊿FliC) in liquid media for 12 h at 37℃.
- Inoculated again in liquid media upto about OD600=2 at 37℃
- Washed, adjusted the cell density, and mixed them. (Sender:Receiver=1000μL:1000μL)
- Incubated at 30°C, measuring the fluorescent intensity intermittently
|
|
| [http://partsregistry.org/Part:BBa_S03623 BBa_S03623] |
|
|
Status (Ongoing)
- We have constructed all three mutants described above.
- We haven't seen any noticeable difference in response-time between wt-luxR and I45F (hyper-sensitive) mutant of LuxR. Sucks big time. We think this is probably due to that LuxR is already sensitive enough.
- We have just constructed Leu42 mutants, but they are waiting for the sequence verification (as of Oct 29th). We will conduct the communication experiment as soon as sequencing get completed. Hope we will see the significant time-delay, ideally without any decrease in the endpoint signal intensity.
One more plan; Searching for Delay Switcher
We are planning to do a simple directed evolution to isolate the 'delay switcher' mutant of LuxR.
Reference
- Collins CH et.al. Directed evolution of Vibrio fischeri LuxR for increased sensitivity to a broad spectrum of acyl-homoserine lactones. Mol. Microbiol. 55, 712–723 (2005)
- Koch B et.al.:The LuxR receptor: the sites of interaction with quorum-sensing signals and inhibitors. Microbiol. 2005 Nov;151(Pt 11):3589-602.
- Zhang RG et. al. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA., Nature, 417, 971-974 (2002)
| Home | The Team | The Project | Parts Submitted to the Registry | Notebook |
|---|
 "
"