Team:ETH Zurich/Modeling/Overview
From 2008.igem.org
Luca.Gerosa (Talk | contribs) |
Luca.Gerosa (Talk | contribs) (→Overview on the modelling framework) |
||
| Line 14: | Line 14: | ||
==Overview on the modelling framework== | ==Overview on the modelling framework== | ||
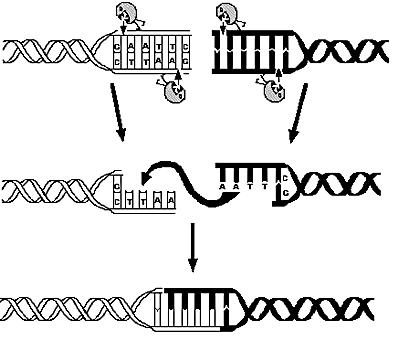
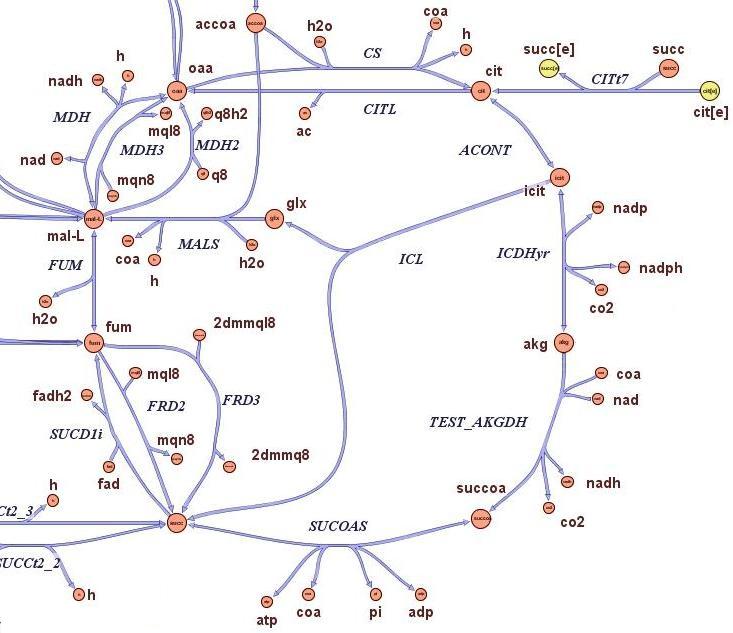
| - | This page is meant to give an introduction to the the overall modelling framework we have constructed in order to asses feasibility analysis, temporal scale details and other parameter estimations that regard our project setup. As introduced in the [[Team:ETH_Zurich/Project/Overview|project overview section]], four main components can be identified in the deviced mechanism. Accordingly, we divided the modelling framework in four modules that tackles the relative problematics. The first module is concerned with the analysis of restriction enzymes and their cutting pattern on E.Coli genome, the second module | + | This page is meant to give an introduction to the the overall modelling framework we have constructed in order to asses feasibility analysis, temporal scale details and other parameter estimations that regard our project setup. As introduced in the [[Team:ETH_Zurich/Project/Overview|project overview section]], four main components can be identified in the deviced mechanism. Accordingly, we divided the modelling framework in four modules that tackles the relative problematics. The first module is concerned with the analysis of restriction enzymes and their cutting pattern on E.Coli genome, the second module predict the cell's response to the selection pressure and its genome reduction from a system point of view (that is, using a genome scale model), the third module adresses issues related to the sensitivity and setting of the chemostat mechanism, the fourth and final module presents the mathematic model of genetic transcriptional circuit used to control restriction enzymes expression. |
is the use of Restryction Enzymes and Ligases as molecular mechanism for reducing the genome, | is the use of Restryction Enzymes and Ligases as molecular mechanism for reducing the genome, | ||
Revision as of 15:02, 11 October 2008
Overview on the modelling frameworkThis page is meant to give an introduction to the the overall modelling framework we have constructed in order to asses feasibility analysis, temporal scale details and other parameter estimations that regard our project setup. As introduced in the project overview section, four main components can be identified in the deviced mechanism. Accordingly, we divided the modelling framework in four modules that tackles the relative problematics. The first module is concerned with the analysis of restriction enzymes and their cutting pattern on E.Coli genome, the second module predict the cell's response to the selection pressure and its genome reduction from a system point of view (that is, using a genome scale model), the third module adresses issues related to the sensitivity and setting of the chemostat mechanism, the fourth and final module presents the mathematic model of genetic transcriptional circuit used to control restriction enzymes expression. is the use of Restryction Enzymes and Ligases as molecular mechanism for reducing the genome,
|
 "
"