Team:ETH Zurich/Wetlab/Chemostat Selection
From 2008.igem.org
(→Growth experiments with E. coli strains of different genomic sizes) |
(→Growth experiments with E. coli strains of different genomic sizes) |
||
| Line 60: | Line 60: | ||
For this we used MG1655 and MDS42, 2 strains with the same background, which differ in genome size by ... % (bp) (link to scarabeus). | For this we used MG1655 and MDS42, 2 strains with the same background, which differ in genome size by ... % (bp) (link to scarabeus). | ||
For a first characterisation of the growth of these 2 strains, we performed growth experiments in LB. | For a first characterisation of the growth of these 2 strains, we performed growth experiments in LB. | ||
| + | |||
| + | |||
[graph with growth curve MG1655 and MDS42 in LB, experiment 30 juli or experiment 10.9.08] | [graph with growth curve MG1655 and MDS42 in LB, experiment 30 juli or experiment 10.9.08] | ||
| - | + | ||
We observed a higher growth rate for MG1655 (mu =) than for MDS42 (mu =). | We observed a higher growth rate for MG1655 (mu =) than for MDS42 (mu =). | ||
Revision as of 14:53, 29 October 2008
|
Chemostat selectionGoalThe goal of our project is to find the minimal genome that is able to create a living E. coli in a specific environmental condition. In the previous section, we have introduced the method we want to apply for reducing the genome. However, manually selecting for those bacteria that have successfully reduced their genomes is obviously impossible. Therefore, we have to find a mechanism that automatically selects cells possessing a reduced genome. IdeaOur idea is to set up a continuous bacterial culture in which cells with a reduced genome overgrow those maintaining more chromosomal DNA. However, bacteria with a smaller genome do not automatically grow faster than those containing more DNA. This is due to the fact that the rate of proliferation is not only influenced by the replication velocity, but also by other factors such as protein synthesis, etc. Therefore, we have to introduce a constraint that renders DNA synthesis the limiting factor of the growth rate. In this scenario, reducing chromosomal DNA would speed up replication leading to fastest growth of those cells with the smallest genome. This way, we would not have to look for bacteria with reduced genomes, but we could let them do it for us! MethodChemostatA chemostat is the instrument of choice for setting up a continuous culture. It is characterized by a continuous influx of medium, and an efflux of bacterial culture of the same volume. Since the volume of the continuous culture is kept constant, the growth rate of the population can be set by the dilution rate. Under these conditions, slowly growing (growth rate < dilution rate) cells will be washed out of the culture, while the fastest growing clone will take the lead and finally dominate the entire culture. A model of the selection mechanism can be found in Chemostat Selection, where we estimate the initial parameters for optimal selection.
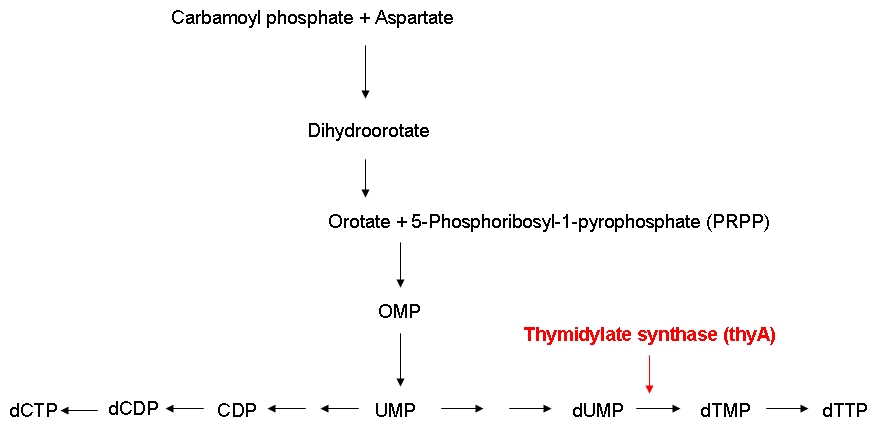
Growth constraintAs mentioned above, we want to introduce a constraint that renders DNA synthesis the limiting factor of the growth rate. DNA synthesis in bacterial cells is accomplished using the following biosynthetic pathway:
The thymidylate synthase catalyzes the methylation of dUMP to yield dTMP. Phosphorylation then converts dTMP into dTTP, one of the four building blocks of DNA. In contrast to the other three nucleotides DNA is made of, dTTP is exclusively used for DNA, but not for RNA synthesis. Therefore, impairing the activity of the thymidylate synthase should interfere quite specifically with DNA synthesis.
The underlying assumption is that a thymidylate synthase knockout can no longer synthesize DNA from glucose and thus depends entirely on the availability of thymidine in the medium. Therefore, if the thymidine concentration is kept low, DNA replication should be slowed down. A decreased replication rate then results in a slower growth rate. This way, by fixing the thymidine supply rate, we can influence the growth rate of the knockout mutant. Supporting our assumptions, Escartin et al. showed that an E. coli strain expressing an enzymatically less active thymidylate synthase shows a significantly lower incorporation rate of radioactively labeled thymidine into DNA indicating a reduced replication velocity (1). The same strain grows poorly in thymidine-deprived growth media. Other reports support that a decreased replication rate results in a slower growth rate and decreased replicative fitness (2). Lab resultsGrowth experiments with E. coli strains of different genomic sizesTo test our hypothesis of selecting for strains with smaller genome in a chemostat situation, we have chosen, to compare growth rate and thymidine yield in strains with different genome size. For this we used MG1655 and MDS42, 2 strains with the same background, which differ in genome size by ... % (bp) (link to scarabeus). For a first characterisation of the growth of these 2 strains, we performed growth experiments in LB.
Labeling of E. coli strains of different genomic sizesThymidylate synthase knockout using phage transductionGrowth experiments with thymidylate synthase knockout strainsKeio thymidylate synthase knockout strainsPhage transduction knockout strainsReferences(1) Escartin F., Skouloubris S., Liebl U., Myllykallio H. (2008): Flavin-dependent thymidylate synthase X limits chromosomal DNA replication. Proc Natl Acad Sci 22 105(29):9948-52. (2) Helmstetter C. (1996): Timing of synthetic activities in the cell cycle. Escherichia coli and Salmonella: Cellular and Molecular Biology. eds Neidhardt FC et al. (Am Soc Microbiol, Washington D. C.): 1627–1639. |
 "
"