Team:Edinburgh/Modelling/Kinetic
From 2008.igem.org
Contents |
Introduction
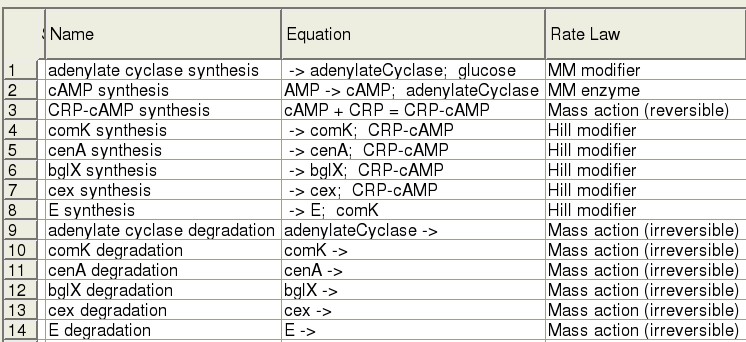
This section aims to outline the model formulation of the secretion pathway for cellulose degradation enzymes as constructed by the University of Edinburgh 2008 iGEM team. The pathway includes ..., The computational model includes two operons, 14 species, and 15 reactions, and focusses on the switching on of genes for cellulose degradation by CRP.
Kinetic Modelling
Through mathematical modelling of cellular processes, we aim to reflect biological function in sets of equations supplied with parameter values that best fit observed reality (Drillon, G. 2008). If the outcomes of model-based simulations closely correspond to experimental results, this indicates that the underlying biological mechanism may be understood. Through the process of modelling, we may generate new hypotheses, as well as investigating doubts on previous suggestions, or corroborating them [Baralla, 2008]. Modelling in preparation and in response to wetlab work can save money and time by focussing on more likely hypotheses.
A standard way of modelling (non-spatial) physical systems which has been successfully applied to simulate metabolic systems [Bakker et al., 1997] is the use of Ordinary Differential Equations (ODEs). The aim "of using a set of ODEs is to describe a biological phenomenon in order to analyze the time dynamics of the system or to calculate the steady state values of the variables involved in the model."and includes the effects of transcriptional regulation as well as of the changes in concentration of metabolites via enzymes.
"The process of modelling tries to identify the variables and determine the relationships among them. The simulation (the execution of the model), allows to get results from the model. In case of differential equations models, the simulation refers simply to a numerical integration finding a solution to the set of equations. The goal of using a set of ODEs is to describe a biological phenomenon (like the nitrogen assimilation) in order to analyze the time dynamics of the system or to calculate the steady state values of the variables involved in the The goal of using a set of ODEs is to describe a biological phenomenon (like the nitrogen assimilation) in order to analyze the time dynamics of the system or to calculate the steady state values of the variables involved in the model."model."
include the effects of gene regulation as well as the concentration
changes of metabolites via enzymes.
The Biological System
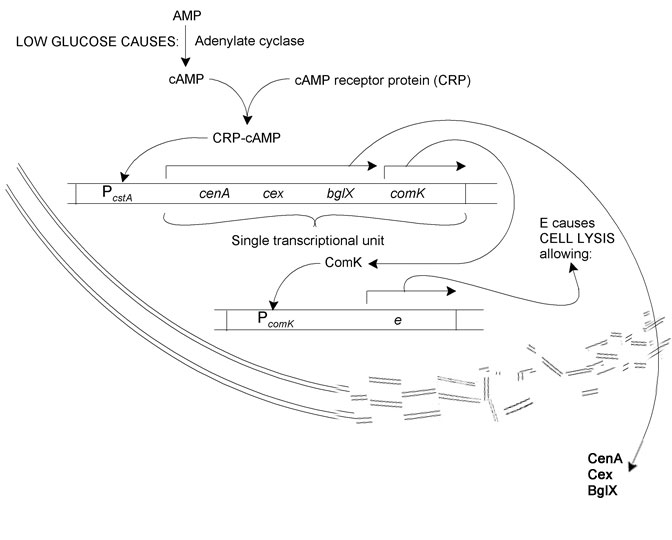
The section we're most interested in modelling is the cellulase production leading to cell lysis pathway.
The system we sought to model is as follows: Reduction in glucose concentration (due to?) leads to increased adenylate cyclase activity. This results in more AMP being converted to cAMP. cAMP binds to CRP. The resultant CRP-cAMP transcription factor binds to the *cstA* operator, causing the transcription of cellulase proteins (cenA, cenB, cenC, and cex) and *comK*. ComK binds to the *comK* operator, whereupon *phiX174E*, a lysis enzyme, is produced. Cell lyses releases the cellulases into the growth medium.
We chose to model the lysis pathway for the cellulose degradation
enzymes, since the activation of the associated genes may be the most
interesting focus for modelling in our system of interest.
cellobiose and beta-glucosidase... hydrolyses the cellobiose to glucose' (O'Neill et al, Gene 44:325-330).
In the absence of experimental measurements, we have to fit likely numbers... Our survey of the literature yielded some kinetic information for the genes in our system, in vivo, as well as in E. coli.
The Model
ODEs
 "
"