Team:Edinburgh/Results
From 2008.igem.org
(Difference between revisions)
| (27 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
==Cellulolysis device== | ==Cellulolysis device== | ||
[[Image:Edinburgh Map1 Cellulolysis.jpg|150px]] | [[Image:Edinburgh Map1 Cellulolysis.jpg|150px]] | ||
| - | * '''''cenA''''' (endoglucanase) | + | * '''''cenA''''' (endoglucanase), '''''cex''''' (exoglucanase) and '''''bglX''''' (beta glucosidase): BioBricks<sup>TM</sup> made and ribosome binding site added; ready to test. Last minute result: possible ''cenA'' and ''bglX'' activity detected using chromogenic substrate, but needs to be repeated and confirmed: hope for more results in our poster and talk at the Jamboree! |
| - | + | * '''P<sub>''cstA''</sub>''' (promoter): BioBrick<sup>TM</sup> made and tested - successful. | |
| - | + | ** [[Team:Edinbrugh/Results/PcstA-xylE|P<sub>''cstA''</sub> promoter assay with ''xylE'' as reporter]] | |
| - | ** [[Team:Edinbrugh/Results/PcstA-xylE| | + | |
== Glycogenesis device == | == Glycogenesis device == | ||
[[Image:Edinburgh Map2 Glycogen.jpg|150px]] | [[Image:Edinburgh Map2 Glycogen.jpg|150px]] | ||
| - | * '''''glgC''''' and '''''glgC16''''' (ADP-glucose pyrophosphorylase, responsible for rate limiting step in glycogenesis): | + | * '''''glgC''''' and '''''glgC16''''' (ADP-glucose pyrophosphorylase, responsible for rate limiting step in glycogenesis): BioBricks<sup>TM</sup> made and tested - successful. |
| - | ** [[Team:Edinburgh/Results/Glycogen1|Glycogen Assay 1 (Qualitative)]] | + | ** [[Team:Edinburgh/Results/Glycogen1|Glycogen Assay 1 (Qualitative: Iodine)]] |
| - | ** [[Team:Edinburgh/Results/Glycogen2|Glycogen Assay 2 (Qualitative)]] | + | ** [[Team:Edinburgh/Results/Glycogen2|Glycogen Assay 2 (Qualitative: Iodine)]] |
| - | ** [[Team:Edinburgh/Results/Glycogen3|Glycogen Assay 3 (Quantitative)]] | + | ** [[Team:Edinburgh/Results/Glycogen3|Glycogen Assay 3 (Quantitative: Raman Spectroscopy)]] |
| + | ** [[Team:Edinburgh/Results/Glycogen4|Glycogen Assay 4 (Quantitative: Raman Spectroscopy)]] | ||
== Starch synthesis device == | == Starch synthesis device == | ||
[[Image:Edinburgh Map3 Starch.jpg|150px]] | [[Image:Edinburgh Map3 Starch.jpg|150px]] | ||
| - | * ''''' | + | * '''''SU1''''' and '''''ISO2''''' (isoamylases): Work in progress - pre-BioBrick<sup>TM</sup> constructs in pSB1A2 have been made using the BABEL protocol, but it is still necessary to remove internal EcoRI sites before fully compliant BioBricks<sup>TM</sup> will be ready to test. This work will be continued in an undergraduate honours project. |
==Lycopene generator == | ==Lycopene generator == | ||
[[Image:Edinburgh Map4 BCarotene.jpg|150px]] | [[Image:Edinburgh Map4 BCarotene.jpg|150px]] | ||
| - | * '''''dxs+crtE+crtB+crtI+crtY''''': Work in progress - encountered problem at final step. | + | * '''''dxs+crtE+crtB+crtI+crtY''''': Work in progress - encountered problem at final step. Assembly of the three genes ''crtE, crtB'' and ''crtI'' together in one construct has failed so consistently and in so many unexpected ways that we are starting to wonder whether this combination is toxic to our host strain under the growth conditions used, though there seems no obvious reason to expect this. Further experiments are required. |
==Limonene synthase device== | ==Limonene synthase device== | ||
[[Image:Edinburgh Map5 Limonene.jpg|150px]] | [[Image:Edinburgh Map5 Limonene.jpg|150px]] | ||
| - | * '''''dxs+ | + | * '''''dxs''+''LIMS1''+''appY''''': BioBrick<sup>TM</sup> made - still being tested. A GC/MS test of an ethyl acetate extract from an induced culture did not detect limonene, but the extraction procedure still needs to be properly validated, so we still can't be sure whether the limonene synthase is working in our host. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 02:00, 30 October 2008
Results and current status as of 29 October 2008. We are hoping to obtain some last-minute results to present at the Jamboree.
Contents |
Overview
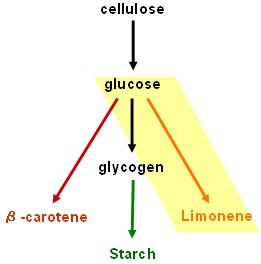
Our results are promising but our dream of using bacteria to convert cellulose into starch and β-carotene is still unrealised. Fortunately, the work will be continued by two undergraduate students after Christmas.
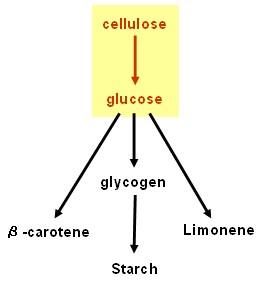
Cellulolysis device
- cenA (endoglucanase), cex (exoglucanase) and bglX (beta glucosidase): BioBricksTM made and ribosome binding site added; ready to test. Last minute result: possible cenA and bglX activity detected using chromogenic substrate, but needs to be repeated and confirmed: hope for more results in our poster and talk at the Jamboree!
- PcstA (promoter): BioBrickTM made and tested - successful.
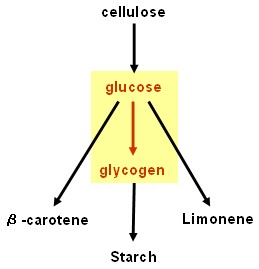
Glycogenesis device
- glgC and glgC16 (ADP-glucose pyrophosphorylase, responsible for rate limiting step in glycogenesis): BioBricksTM made and tested - successful.
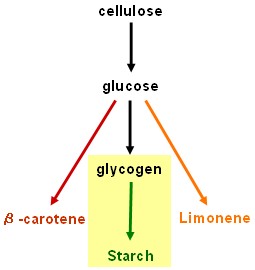
Starch synthesis device
- SU1 and ISO2 (isoamylases): Work in progress - pre-BioBrickTM constructs in pSB1A2 have been made using the BABEL protocol, but it is still necessary to remove internal EcoRI sites before fully compliant BioBricksTM will be ready to test. This work will be continued in an undergraduate honours project.
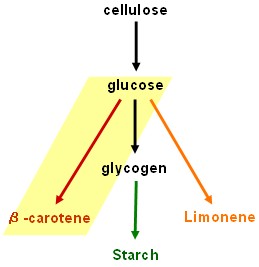
Lycopene generator
- dxs+crtE+crtB+crtI+crtY: Work in progress - encountered problem at final step. Assembly of the three genes crtE, crtB and crtI together in one construct has failed so consistently and in so many unexpected ways that we are starting to wonder whether this combination is toxic to our host strain under the growth conditions used, though there seems no obvious reason to expect this. Further experiments are required.
Limonene synthase device
- dxs+LIMS1+appY: BioBrickTM made - still being tested. A GC/MS test of an ethyl acetate extract from an induced culture did not detect limonene, but the extraction procedure still needs to be properly validated, so we still can't be sure whether the limonene synthase is working in our host.
 "
"