Team:Groningen/Modeling
From 2008.igem.org
(Difference between revisions)
| Line 2: | Line 2: | ||
<div style="width: 700px; margin-left: 15px; float:right;"> <html><!-- main content div --></html> | <div style="width: 700px; margin-left: 15px; float:right;"> <html><!-- main content div --></html> | ||
To gain insight into the working of our designs we try to develop reliable and realistic models. | To gain insight into the working of our designs we try to develop reliable and realistic models. | ||
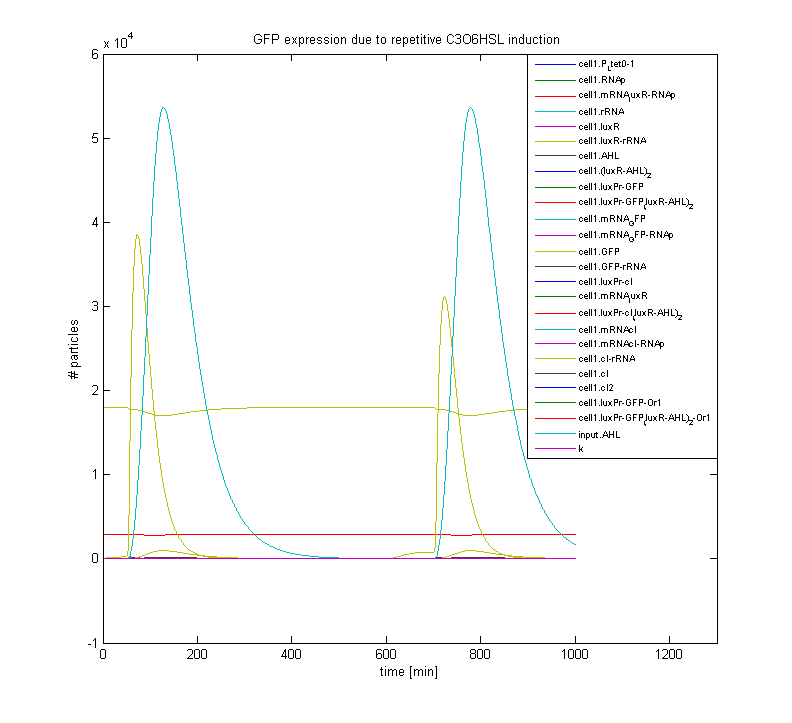
| - | + | The ODE solvers incorporated in the SimBiology package, for Mathworks' Matlab, are employed to model the genetic design on the single cell level, displaying the variations in concentrations of all relevant substances over time. | |
| - | + | To examine the workings of our designs on the multi-cell level we are developping a routine in Matlab which can predict the the collective behaviour of the designed cells on a 2D grid. Hopefully this will help us to gain more insight in the characteristics of AHL exchange between the cells. | |
[[Image:figforsite.png|left|frame|400px|Academic building in Groningen]] | [[Image:figforsite.png|left|frame|400px|Academic building in Groningen]] | ||
Revision as of 12:33, 29 July 2008
To gain insight into the working of our designs we try to develop reliable and realistic models. The ODE solvers incorporated in the SimBiology package, for Mathworks' Matlab, are employed to model the genetic design on the single cell level, displaying the variations in concentrations of all relevant substances over time. To examine the workings of our designs on the multi-cell level we are developping a routine in Matlab which can predict the the collective behaviour of the designed cells on a 2D grid. Hopefully this will help us to gain more insight in the characteristics of AHL exchange between the cells.
 "
"