Team:Guelph/Project
From 2008.igem.org
| Home | The Team | The Project | Parts | Notebook | Modeling | Links |
|---|
Contents |
Project Abstract
To produce probiotic and endophytic microbes with nutrient or RNAi inducing capabilities, we made two Biobricks.... ...We tested said microbes in vivo and monitored their activity by GFP expression. Our results were X mg of betacarotene production by lactobacillus y in our intestinal model and complete silencing of target plant genes. ---> as abstracts summarize a completed project, this will be completed when our lab work is done.
Overall project
Microbes are found in every nook and cranny of the planet, and multicellular organisms are no exception. Plants and animals are found to contain huge numbers of bacteria and fungi that help with nutrient absorption, producing beneficial compounds, fighting off pathogens, or often are pathogens themselves. We are interested in taking advantage of some of these microbes to deliver transgenic payloads for the benefit or modification of the host organism. These might be called GM endosymbionts. On the human side, we would like to introduce the carotendoid metabolic genes from a well studied soil microbe called Erwinia urodevora into human probiotic microbes which will survive and colonize the intestine for stable production of the essential human nutrient, pro-vitamin A. Time permitting, we will attempt to enhance carotenoid accumulation by increasing plasma membrane sink by overexpression of the fumarate reductase operon.
Millions of humans suffer from vitamin A deficiencies across the world, resulting in blindness and death which could be mitigated by symbitic production of this important vitamin. We will also attempt to get corn endosymbionts to produce pro-vitamin A, a function usually performed by the plant itself. In both cases, the ethical and practical implications of this technology will affect how it might ever be implemented, so we are very excited to be collaborating with the Calgary Ethics team to ruminate on the possibilities and pitfalls of this bit of synthetic biology.
A more basic wet lab project we are doing will focus on RNAi signal delivery by a corn plant endosymbiont to silence corn genes. Most euaryotes react to double stranded RNA by copying it and chopping up any mRNA with the same sequence, while prokaryotes do not seem to possess the same response. Since bacteria like K. pneumonii live in large stable populations within corn plants, it is believed that as individual bacteria grow, die and lyse within the plant host, they will release RNAi transcripts into the sensitive host during the entire life cycle of the plant, which will silence the targeted gene and show a phenotype indicating gene function. Bacterial Induced Gene Silencing (BIGS) will be a useful, quick, and stable alternative for plant functional genomic research. This will be an enableing technology for scientists studying plant gene function, as well as a Biobrick registry value adding tool; any Biobrick compliant peice of DNA will be clonable into a double stranded RNA orientation that could be expressed by bacteria (or plants for that matter).
Project Details
Background and Justification
1) Beta-carotene production by endosymbionts: For socioeconomic and environmental reasons, this might be advantageous. To a large extent, this will be answered by our collaborators at iGEM Calgary Ethics, so stay posted for their results and paper.
2) BIGS for functional genomics: Functional genomics is an area of genetics that uses genomic information to help understand gene function. Currently, the most straightforward way to do this for any given gene is to analyze a single gene mutant to discover that organism's deficiency, and thus the gene's normal function. If you're lucky, you can find a natural mutant, or make one using random mutagenisis. However, in complex eukaryotes with tens of thousands of genes, it is impossible to expect that you can get knock outs for any particular gene you want to study. As of a decade ago, it began to be possible to use RNAi to selectively knock out your particular gene by making a transgenic organism expressing double stranded RNA with the same sequence of the target gene. For a eukaryote like corn, this makes it possible knock out one particular gene you want to study, but it does require about a year for that transgenic corn to be made. An alternative in some eukaryotes is Viral Induced Gene Silencing, or VIGS, whereby the double stranded RNA is transiently delivered into the organism by a virus. VIGS has only been developed for certain eukaryotes however (not including corn), and even so is only a transient way to silence a gene, giving a scientist only a portion of the eukaryote's life cycle where gene function might be studied. We ask ourselves, can a system be made which could effectively silence eukaryotic genes using RNAi, but more quickly and cheaply than making a transgenic eukaryote and more stably and long term than by using VIGS? Our idea is that a normal endosymbiont of an organism could do this if it can be coaxed to produce this RNAi signal. Read on to find out how we'll test this idea...
The Experiments
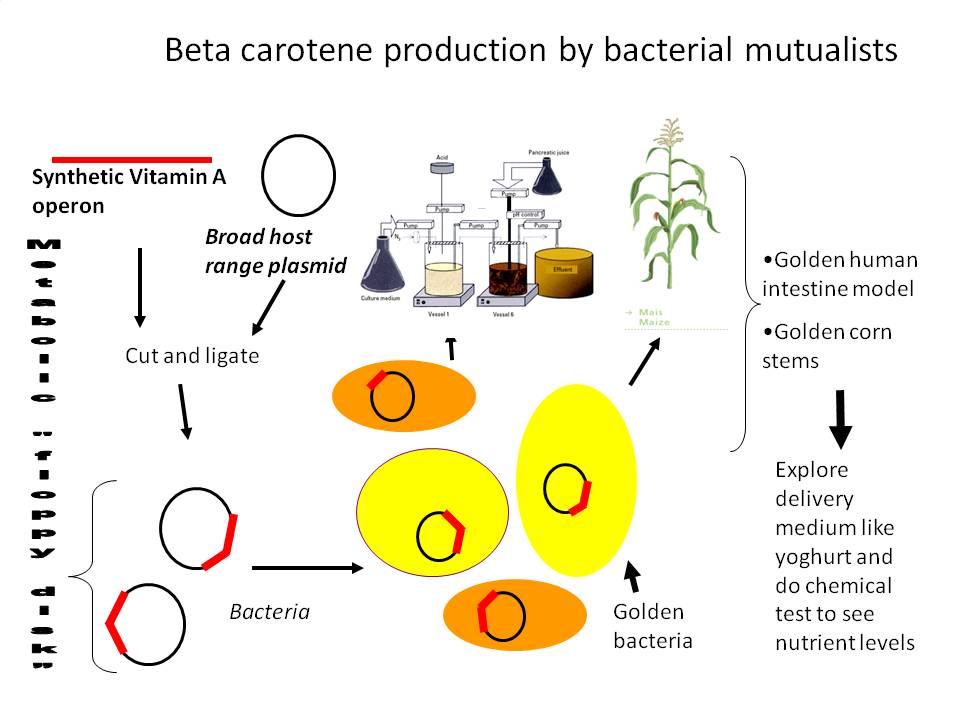
1) Beta carotene producing intestinal microbes and plant endophytes
Tentative cloning strategy:
So what is the stategy we should follow? Initially this Hydrogen synthetic operon paper was inspiring us:
http://www.ncbi.nlm.nih.gov/pubmed/17996187
Its specific for a bacterial synthetic operon and if we followed it we wouldn't have to rework things too hard in order to follow it.
However, this was not a perfectly Biobrick compliant strategy, and there were unecessary extra steps we'd have to follow so we reworked our strategy. Now we will put into a Biobrick vector a strong constitutive promoter from herbicide tolerant Amaranthus weeds which is unregulated (and therefore constitutive) in prokaryotes. To do this we should PCR up the promoter from the pDSK-GFPuv plasmid (introducing an SpeI site after the native NdeI), and put it into this:
http://partsregistry.org/wiki/index.php?title=Part:pSB1AK3
Likewise, we are PCRing up and inserting a restriction site optimized GFP into the plasmid. Then, we're ready to clone the carotenoid genes into the NdeI space in between promoter and GFP.
The gene order for the operon has already been optimized. This paper explains we should have E-B-I-Y. Check it out:
http://aem.asm.org/cgi/content/abstract/AEM.02268-06v1
The initial gene, crt-E, on plasmid p3-10-10 will be amplified and appropriate flanking NdeI and XbaI restriction sites introduced, as well as a SpeI and an XhoI site after the stop codon of the gene. There is also a ribosome binding site present in front of each of the carotenoid genes, and thus we will conserve them in our cloning strategy. We will cut the plasmid with NdeI and SpeI, and the PCR product with NdeI and XbaI. SpeI and Xba I being sticky-end cutters with complementary overhangs, their respective cut ends will ligate to each other. This will destroy the SpeI site on the plasmid and the XbaI site on the PCR product, but these complementary sticky ends have the benefit of decreasing the rate of background ligations; the complementary sticky ends act as 'heat seeking missiles', seeking out their complementary mates and ignoring non-complementary overhangs or blunt end.
The crt-B PCR product is flanked with XbaI and XhoI, with an internal SpeI site after the stop codon of the crt-B gene. To insert the crt-B after crt-E, we will cut it with XbaI and XhoI, while cutting the plasmid with SpeI and XhoI. The complementary pairing between SpeI and XbaI's sticky ends will again allow them to ligate, destroying the Spe and Xba sites in the process.
Next comes Crt-I. This is done in the same way as above, as it is also flanked with XbaI and XhoI with an internal SpeI site. Once again, the PCR product is cut with XbaI and XhoI, while the plasmid is cut with SpeI and XhoI. Ligate.
(note: Crt-I has two PstI sites which really should be removed but don't have to if we don't have time - I have a technique I'd like to try to do this using some sort of nested PCR - there are a couple primers in the list which could be used to use to do this)
Finally, we insert Crt-Y. Cut it (PCR product) with XbaI and XhoI, while cutting the plasmid with SpeI and XhoI. Ligate.
The GFP at the end should hopefully work as a reporter to let us know the transcript is OK, and the RBS sites are all lined up properly. If we want to add any more genes, we use the same protocol with XbaI, SpeI and XhoI. One candidate extra addition to the operon could be fumarate reductase since its overexpression results in increased cell membrane, which is where lipid soluble carotenoids are stored. Here's the link the paper:
http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=215469
In plants it has been shown that sink strength is a limiting factor to carotenoid accumulation, so giving bacteria more carotenoid storage space may result in much more carotenoid accumulation. Here's a discussion about engineering plant sinks for increased carotenoid accumulation:
http://www.isb.vt.edu/articles/feb0802.htm
Assay for beta-carotene production.
Transfer to gram positive and gram negative plasmids.
Electroporate into different probiotic strains.
Monitor beta carotene production and GFP in biological system/model.
Celebrate a succesful new technology for improved human health!
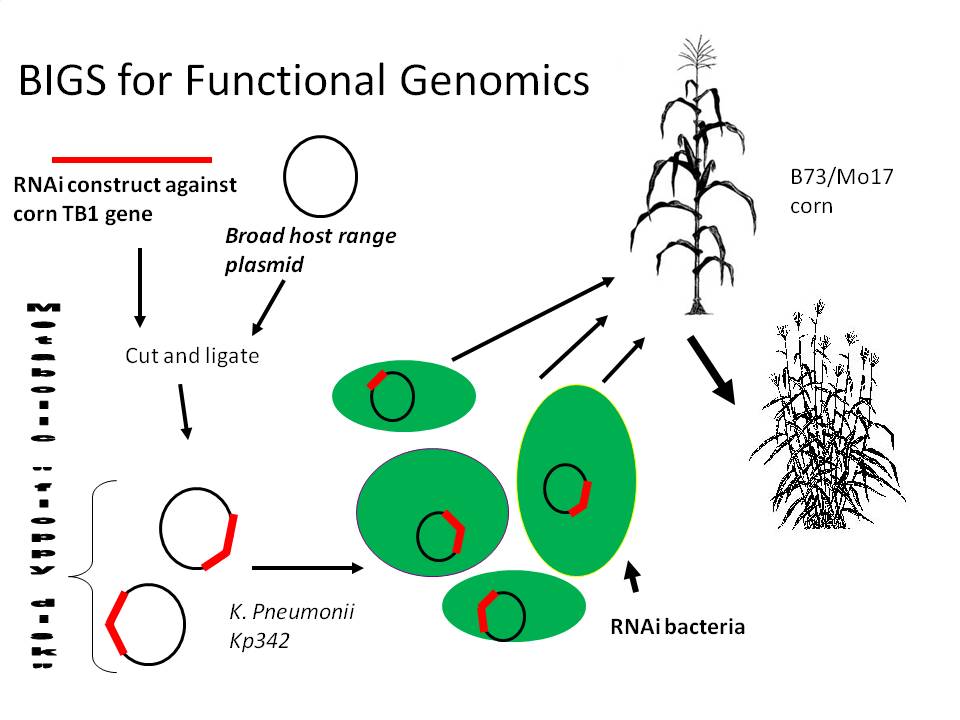
2) RNAi inducing corn endophytes (BIGS)
Functional genomics is the study of gene function in an organism through (genomics) informed study of transcription, translation, interactions, and phenotype. A common way to do this is by making mutants and assaying for different phenotypes; a very convenient way to accomplish this is by using RNA interferance (RNAi). Usually this is done by making transgenic plants expressing RNAi constructs, and is stable but limited by the difficulty and length of time it takes to produce the transgenic plant. In the case of corn, this means almost a full year before you can test your gene function.
An alternative is called viral induced gene silencing (VIGS) which can uses a plant specific virus to deliver and transiently silence the gene. An obvious limitation here is that it is a transient process even if it has been developed for a particular plant, which it has not been for corn.
We propose a happy medium which would be relatively easy and cheap, stable during the whole lifecycle of the plant, Biobrick compatible (allowing RNAi constructs to be made from any biobrick compatible fragment) and importantly, quick and effective. Bacterial induced gene silencing (BIGS) will employ an endophytic microbe (lives mutualistically inside a plant) to deliver an RNAi signal against a plant gene in order to study that gene's function. We are hoping that as these microbes die, they will release these molecules into the medium, and since plants are very sensitive to these double stranded RNA molecules, they will sense AND systemically amplify the signal across the entire plant body.
There are relevant precedants for this area of research, but it has never before been done using endophytes in plants which can produce this signal during the entire life cycle of the host. Here are some relevant publications supporting the potential of our work which we will not further discuss:
Spraying RNAi E. Coli onto plants to protect against viruses - http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=153545
Using E. coli to deliver RNAi signals to tumors in mouse colons - http://www.ncbi.nlm.nih.gov/books/bv.fcgi?indexed=google&rid=eurekah.section.75458
We are planning to order a 200 bp Zea mays actin2 intron construct from our sponsoring DNA synthesis company, IDT, which will consist of shuffled Biobrick ends (XbaI and PstI will switch spots) which will allow RNAi compatibility with any pre-existing Biobrick in the registry. We will program the ribosome binding site sequence AGGAGG into the middle of the intron, as well as the 3' and 5' ends of the construct to improve RNA stability by ribosome binding. We will also program NdeI and XhoI restriction sites into the intron where we will clone a GFP gene for monitoring of RNAi construct transcription in vivo. This construct will be ligated to the strong termination signal found in the Biobrick part found here: http://partsregistry.org/wiki/index.php?title=Part:BBa_B1006 , ligated to the strong constitutive PbsA promotor, and then sense and antisense version of either a GFP or corn gene will be inserted into the RNAi Biobrick polylinkers. GFP silencing will be tested by innoculation of Arabidopsis plants constitutively expressing GFP, while corn gene silencing activity will be monitored by testing endophyte silencing efficiency of the TB1 gene whose expression inhibits branch formation in corn plants.
 "
"