Team:HKUSTers/Initial Proposal/Randomizer
From 2008.igem.org
(Difference between revisions)
(New page: __NOTOC__ <html> <head> <style> .ideasList{ margin-bottom:30px } ul#sub { margin:0px; padding:0px; font-size:14px; list-style-image:none; list-style-type:none; } #sub li { margin...) |
|||
| Line 128: | Line 128: | ||
</table> | </table> | ||
</html> | </html> | ||
| + | |||
| + | |||
| + | ==The core circuit design== | ||
| + | Two promoters locate adjacent to each other, so that the first RNA polymerase binding event could be random if two promoters have similar binding affinity. And this will generate establishing effect; one gene products will have higher copy number, which could effectively repress the neighbor promoter, and further induce its own expression, thus the optimal result is that either Blue or Red gene is dominated. | ||
| + | |||
| + | [[Image:Promoter 1.png]] | ||
| + | |||
| + | ==Background Information== | ||
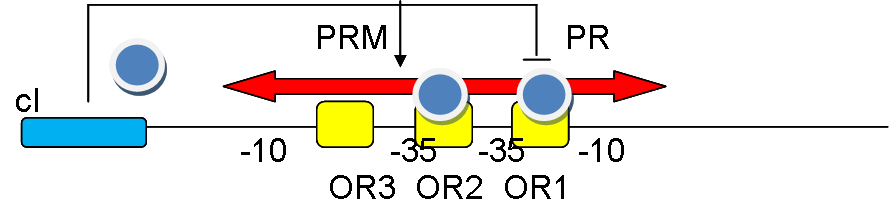
| + | -35 and -10 are important sites for polymerase binding, cI protein is generated by cI gene, which firstly binds to OR1, repress PR, and cooperatively bind to OR2 induce PRM thus its own production. | ||
| + | [[Image:Promoter.png]] | ||
| + | |||
| + | == | ||
Revision as of 04:22, 30 October 2008
| Home | The Team | The Project | Parts | Modelling | Notebook | Gallery |
The core circuit design
Two promoters locate adjacent to each other, so that the first RNA polymerase binding event could be random if two promoters have similar binding affinity. And this will generate establishing effect; one gene products will have higher copy number, which could effectively repress the neighbor promoter, and further induce its own expression, thus the optimal result is that either Blue or Red gene is dominated.
Background Information
-35 and -10 are important sites for polymerase binding, cI protein is generated by cI gene, which firstly binds to OR1, repress PR, and cooperatively bind to OR2 induce PRM thus its own production.
==
 "
"
