Team:Harvard/Dailybook/Week9/Chemical and Light
From 2008.igem.org
Contents |
105, mtrB Ligations/transformations 08/17
Parts to ligated came from 8/17
We used a 2:1 molar ratio of insert to vector for all of the ligations. The ligations were all transformed into TOP10 cells.
| Plasmid | Number of colonies |
| P105+P1'BB | 2 |
| P105+P3'BB | 102 |
| P101+P105 | 0 |
| P101+P108 | 0 |
| (mtrB(BB)+P97)+P108 | 20 |
| (mtrB(BB)+P97)+P116 | 4 |
| (mtrB(BB)+P97)+P117 | 13 |
In retrospect, it appears that all the ligations with mtrB are wrong, since the digest was actually wrong.
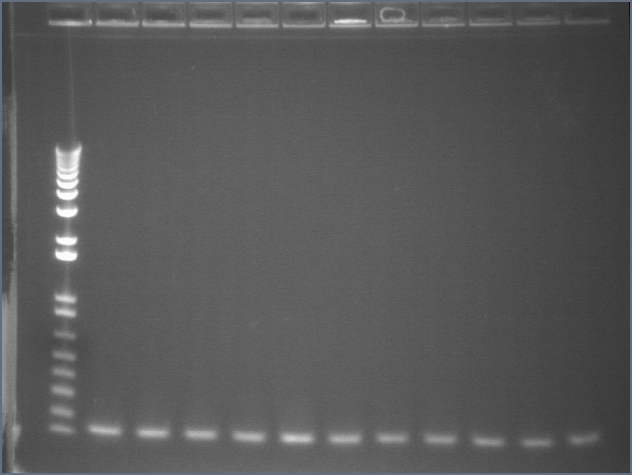
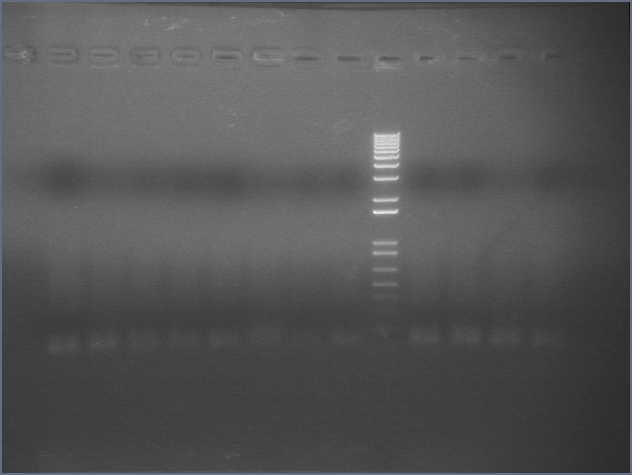
105 Colony PCR 08/18
The annealing temperature was 55 °C and the elongation time was 2'45" (we used Platinum Taq supermix).
Expected band size is 2475bp.
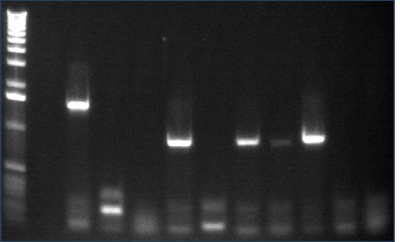
Gel 1: 1-11
- #2, 5 picked for 5mL culture
Lane 1: 1 kb ladder
Lanes 2-3: P105+P1'BB
Lanes 4-12: P105+P3'BB
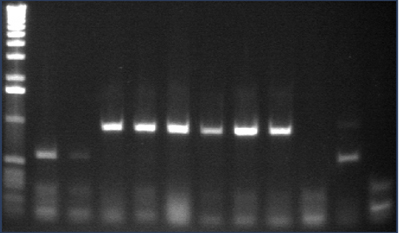
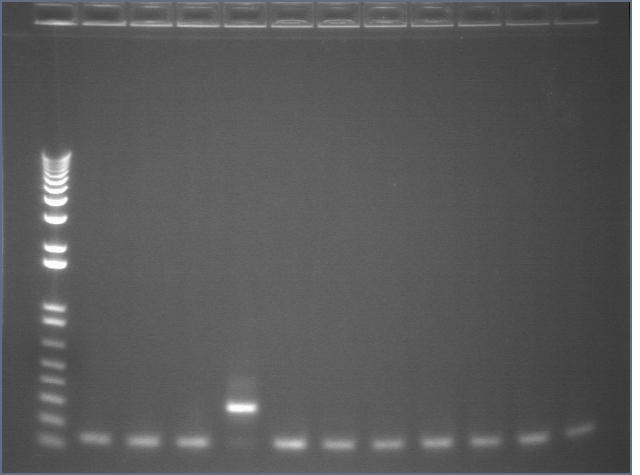
Gel 2: 12-22
- #12, 14 picked for 5mL culture
Lane 1: 1 kb ladder
Lanes 2-12: P105+P3'BB
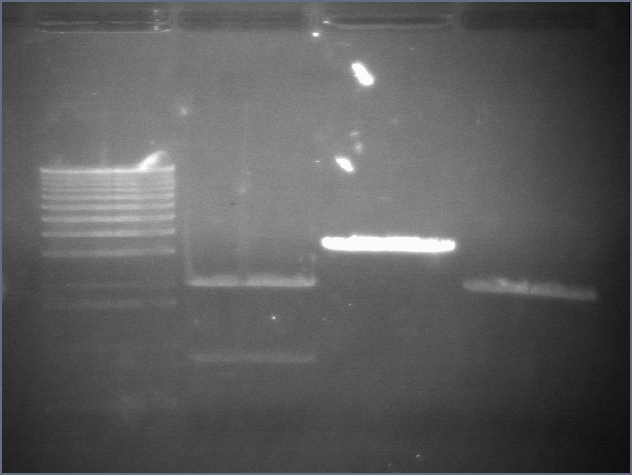
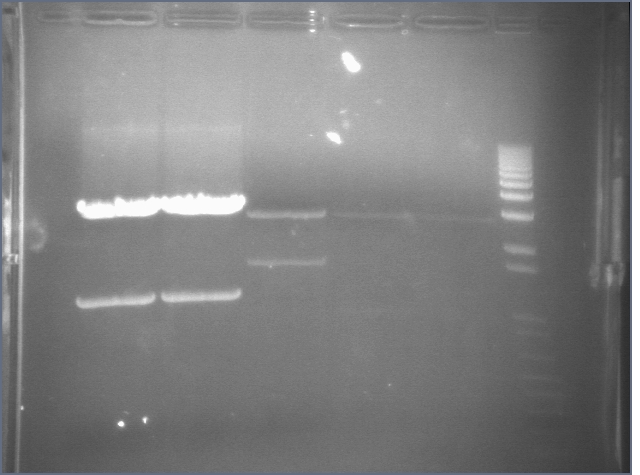
RE digests 45, 63, 97 08/18
We used the Fermentas enzymes and digested for 25'.
Lane 1: 1 kb ladder
Lane 2: P45 cut XP (876; 2079)
Lane 3: P63 cut EX (3284)
Lane 4: P97 cut SP (2091)
QPI+45 Ligations/transformations 08/18
We used the 45 just digested, so all DNA was cut with the Fermentas enzymes.
| Plasmid | Ratio (insert!vector) | Strain | Number of colonies |
| P117? vector control | 2!1 molar with EB | DH5-alpha | 1 |
| P117?+45 | 2!1 molar | TOP10 | 2 |
| P117?+45 | 2!1 molar | DH5-alpha | 30 |
| P116 vector control | 2!1 molar with EB | DH5-alpha | 0 |
| P116+45 | 2!1 molar | TOP10 | 5 |
| P116+45 | 2!1 molar | DH5-alpha | 50 |
| P108 vector control | 2!1 molar with EB | DH5-alpha | 2 |
| P108+45 | 2!1 molar | DH5-alpha | 4 |
| P108+45 | 2!1 molar | TOP10 | 1 |
| P108+45 | 7!1 volumetric | DH5-alpha | 0 |
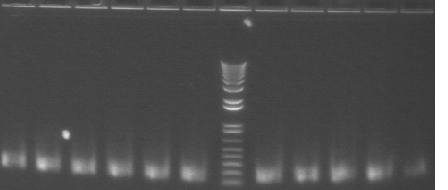
Colony PCR 08/19
The annealing temperature was 55°C and the elongation time was 2'30" for P108+45 and 2' for P116 and P117. We used the Platinum Taq SUPERmix.
Gel 1
Lane 1: 1 kb plus ladder
Lanes 2-5: P108+45 (~2200)
Lanes 6-12: P116+45 (~1800)
Gel 2
Lane 1: 1 kb plus ladder
Lanes 2-12: P116+45 (~1800)
Gel 3
Lane 1: 1 kb plus ladder
Lanes 2-3: P116+45 (~1800)
Lanes 4-12: P117+45 (~1800)
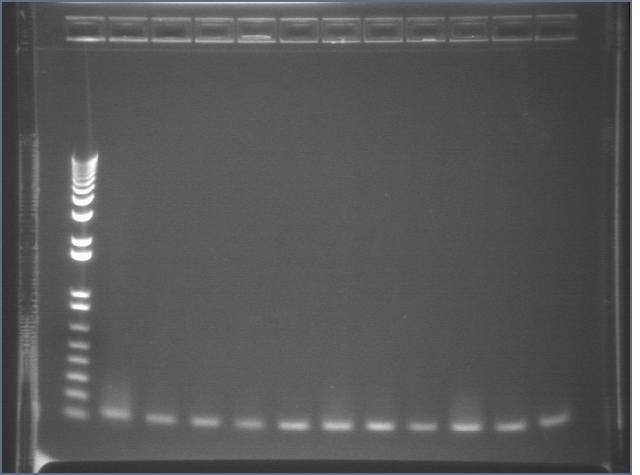
RE digests 101, 115, 118 08/19
Lane 1: P101 cut ES (3221; 1090)
Lane 2: P101 cut XP (3221; 1090)
Lane 3: P118 cut XP (1555; 2750)
Lane 4: P115 cut ES (960; 2750)
Lane 5: 1 kb PLUS ladder
Ligations/transformations of P101+115/118 08/19
We used a 2:1 insert to vector ratio for all ligations. We tried using Amy™'s method of dephosphorylating (i.e. using the Roche kit).
| Plasmid | Strain | Number of colonies |
| P101 vector control (P115) | DH5-alpha | too many to count (same as P101+115 in DH5-alpha) |
| P101+115 | DH5-alpha | too many to count |
| P101+115 | TOP10 | too many to count (more than in P101+115 in DH5-alpha) |
| P101 vector control (P115) | DH5-alpha | too many to count (less than P101+118 in DH5-alpha) |
| P101+115 | DH5-alpha | too many to count |
| P101+115 | TOP10 | too many to count (more than in P101+118 in DH5-alpha) |
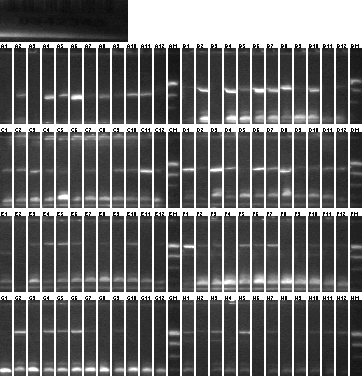
Colony PCR 08/20
We used an annealing temperature of 55 °C and elongation temperature 1'45" and 2'15", and Platinum Taq SUPERmix
Lanes 1-2: P101 (115) vector control
Lanes 3-48: P101+115
Lanes 49-50: P101 (118) vector control
Lanes 51-96: P101+118
- Samples 2, 4, 14, 25, 49, 61, 77 picked for 5mL culture and sequencing.
- It's a bit alarming that the vector controls have ~1kb inserts. Uncut P101 should not transform, as the part contained is lethal (ccdB).
mtrB+RBS PCR 8/20
Rx mix (split into 12):
- 120μL 5x HF buffer
- 12μL 10mM dNTPs
- 15μL each primer
- 6μL resuspended wt S. oneidensis colony
- 6μL Phusion polymerase
- 426μL H2O
Rx conditions
- Initial denaturation: 2:30s @ 98°C
- Denaturations: 10s @ 98°C
- Annealing: 30s @ 55-62°C gradient
- Extension: 45s for first 10 cycles, 55s for next 30 cycles @ 72°C
All the tubes were mysteriously pink after thermocycling.
Attempt 2
Phusion
Rx mix (split into 12):
- 120μL 5x HF buffer
- 12μL 10mM dNTPs
- 15μL each primer
- swirl of wt S. oneidensis colony
- 6μL Phusion polymerase
- 432μL H2O
Rx conditions
- Initial denaturation: 2:30s @ 98°C
- Denaturations: 30s @ 98°C for first 10 cycles, 10s for next 35 cycles
- Annealing: 30s @ 58-64°C gradient
- Extension: 45s for first 10 cycles, 55s for next 35 cycles @ 72°C
Platinum
Rx mix (split into 2):
- 135μL Supermix
- 3μL each primer
- swirl of wt S. oneidensis colony
- 9μL H2O
Rx conditions
- Initial denaturation: 10m @ 95°C
- Denaturations: 45s @ 95°C for first 10 cycles, 30s for next 30 cycles
- Annealing: 30s @ 57°C
- Extension: 2:30s @ 72°C
All conditions failed.
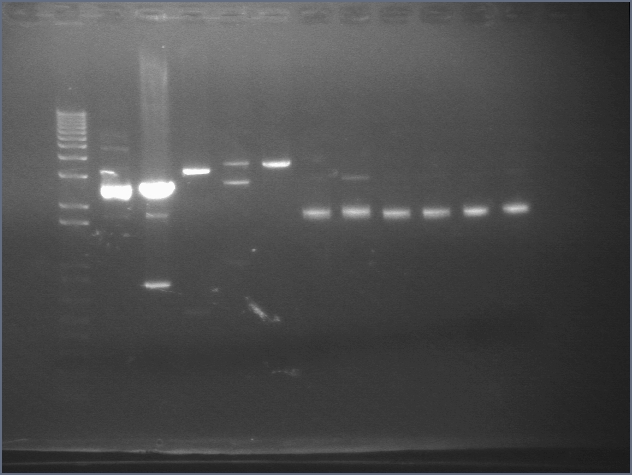
RE digests 08/20
Lane 1: 1 kb plus ladder
Lane 2: P48 uncut (3477)
Lane 3: P48 cut X (3477)
Lane 4: P113 cut XP (534; 3339)
Lane 5: P117 cut SP (3687)
Lane 6: P116 cut SP (3687)
Lane 7: P38+51 (in p15A) B uncut (4155)
Lane 8: P38+51 (in p15A) B cut EP (1405)
Lane 9: P38+51 (in p15A) C uncut (4155)
Lane 10: P38+51 (in p15A) C cut EP (1405)
Lane 11: P38+51 (in p15A) D uncut (4155)
Lane 12: P38+51 (in p15A) D cut EP (1405)
 "
"