Team:Hawaii/Biobrick conversions
From 2008.igem.org
(→Restriction digest) |
(→Restriction digest) |
||
| Line 143: | Line 143: | ||
====Restriction digest==== | ====Restriction digest==== | ||
| - | :* Sequential digest of pRL1383a and | + | :* Sequential digest of pRL1383a and ccdB with HindIII then BamHI in NEBuffer 2 + BSA |
| - | + | ||
| - | + | ||
===Subcloning of lac device and BioBrick MCS in BB-pRL1383a=== | ===Subcloning of lac device and BioBrick MCS in BB-pRL1383a=== | ||
Revision as of 09:52, 31 July 2008
Contents
|
Objectives
- Convert GFP (BBa_E0040) into a fusion brick using site-directed mutagenesis.
- Convert pRL1383a into a Biobrick vector by replacing its natural MCS with the Biobrick MCS
Protocol
PCR mutagenesis of GFP
- Combined:
- 0.5 μl BBa_E0040 plasmid
- 0.5 μl 10mM forward primer
- 0.5 μl 10mM reverse primer
- 3.5 μl nanopure water
- 5 μl Taq (we used EconoTaq Green Taq)
- Ran for 30 cycles of denaturing, annealing, extension
- Initial denature @ 94C for 2 min.
- Denature @ 94C for 30 sec.
- Anneal @ 55C for 30 sec.
- Extend @ 72C for 60 sec.
- Final extension @ 72C for 10 min.
- Held @ 4C inifinitly.
| Primer | Sequence | Length | G/C content | Tm | Notes |
|---|---|---|---|---|---|
| GFP fusion foward | GCCGCTTCTAGAcgtaaaggag | 22 bp | 54.55% | 60.2 C | PCR out from E0040, starts annealing from partial NotI (5 of 8 nucleotides of) site, continues with XbaI, omits TG of ATG codon for site directed mutagenesis, begins again with GFP codon 2-4 (cgt aaa gga) |
| GFP fusion reverse | cgagtcagtgagcgaggaag | 20 bp | 60% | 59.6 C | PCR out from E0040, priming after all end sites (5'taataa t actagt a gcggccg ctgcag gCTTCCTCGCTCACTGACTCG3') |
Extraction of Biobrick MCS
- Combined:
- 0.5 μl BBa_B0034 plasmid
- 0.5 μl 10mM forward primer
- 0.5 μl 10mM reverse primer
- 3.5 μl nanopure water
- 5 μl Taq (we used EconoTaq Green Taq)
- Ran for 30 cycles of denaturing, annealing, extension
- Initial denature @ 94C for 2 min.
- Denature @ 94C for 30 sec.
- Anneal @ 55C for 30 sec.
- Extend @ 72C for 90 sec.
- Final extension @ 72C for 10 min.
- Held @ 4C inifinitly.
| Primer | Sequence | Length | G/C content | Tm | Notes |
|---|---|---|---|---|---|
| HindIII+VF2 | cctAAGCTTtgccacctgacgtctaagaa | 29 bp (20 bp) | 48.3% (50.0%) | 65.9 C (58.6 C) | Includes RE extension HindIII site and three 5' nucleotides for efficient cutting. Parentheses indicate primer information w/o RE site and 3 nucleic acids. Based on VF2 primer. |
| BamHI+VR | ccaGGATCCattaccgcctttgagtgagc | 29 bp (20 bp) | 55.2% (50.0%) | 67.9 C (58.0 C) | Includes RE extension BamHI site and three 5' nucleotides for efficient cutting. Parentheses indicate primer information w/o RE site and 3 nucleic acids. Based on VR primer. |
Subcloning of GFP fusion brick and pRL1383a Biobrick vector
Restriction digest
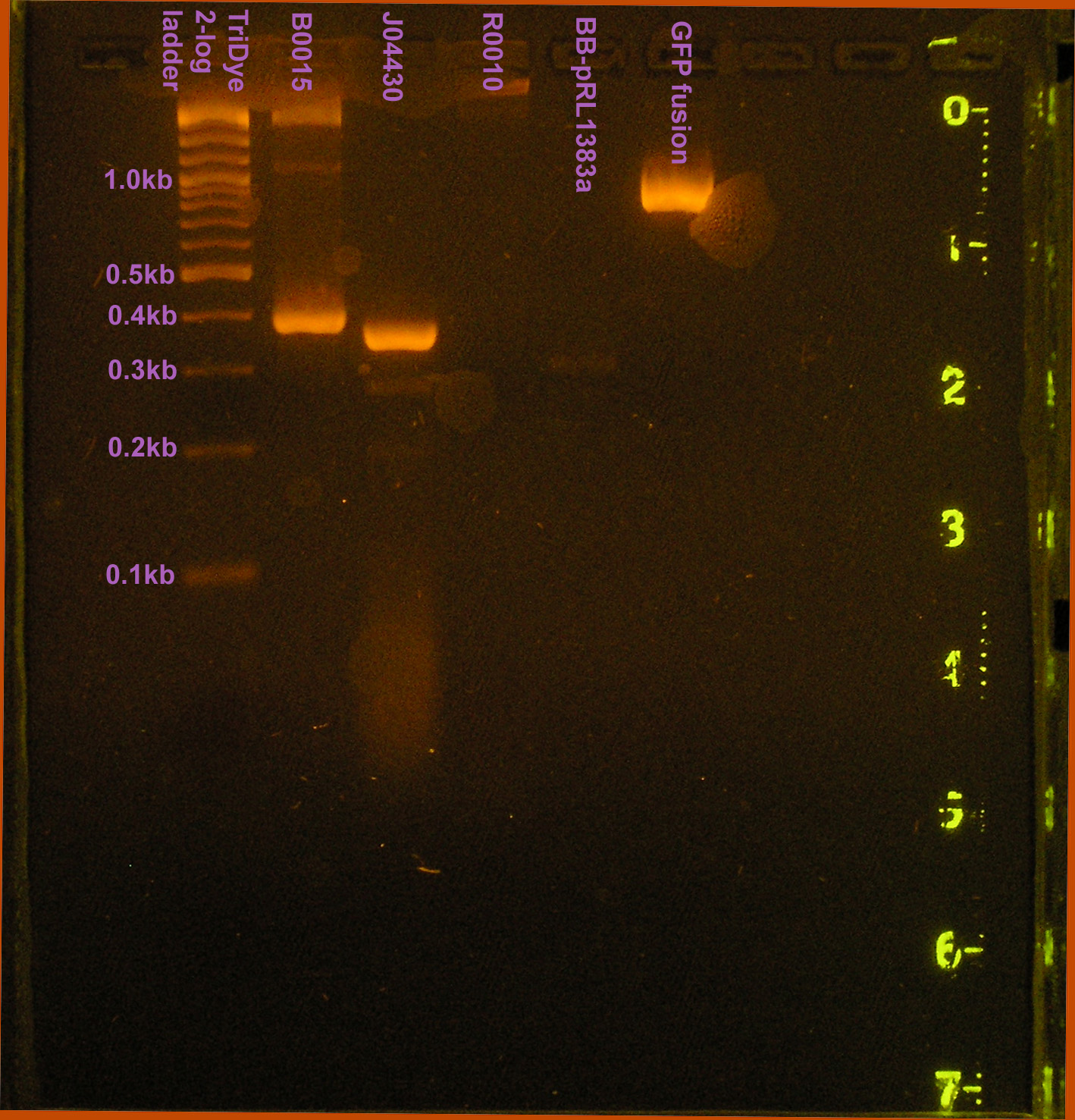
- GFP fusion
- BBa_C0012
- BBa_B0034 derived MCS
- pRL1383a
Ligated insert and vector
- Ran 20 μl ligation reactions
- 7 μl BioBrick segment (~2.8 ng) + 0.5 μl pRL1383a vector (~0.95 ng)
- 7 μl GFP fusion (~7 ng) + 2 μl C0012 derived vector (~1.8 ng)
Transformed into DH5α
- Used 10 μl of ligation reaction to transform.
- Incubated 10 min. on ice after addition of DNA.
- Plated GFP fusion + vector on LB + amp100
- Plated MCS + pRL1383a on LB + sp100
Verification of parts
Colony PCR
Sequencing
- 5 μl of the PCR products were digested with 2 μl ExoSAP.
- Samples were sent to CORE Hawaii for sequencing.
- 12 μl sample with 3.2 pmol primer and 20ng/100bp concentration of PCR product
Reconstruction of BB-pRL1383a
PCR extraction of ccdB gene and BioBrick MCS
- Combined:
- 1.0 μl pSB1A3 plasmid
- 0.5 μl 10mM forward primer
- 0.5 μl 10mM reverse primer
- 3 μl nanopure water
- 5 μl Taq (we used EconoTaq Green Taq)
- Ran for 30 cycles of denaturing, annealing, extension
- Initial denature @ 94C for 2 min.
- Denature @ 94C for 30 sec.
- Anneal @ 54, 58, 62C for 30 sec.
- Extend @ 72C for 90 sec.
- Final extension @ 72C for 10 min.
- Held @ 4C inifinitly.
(Note: same primers as before)
Subcloning of ccdB and BioBrick MCS in BB-pRL1383a
PCR extraction of lac device and BioBrick MCS
- Combined:
- 1.0 μl BBa_J33207
- 0.5 μl 10mM forward primer
- 0.5 μl 10mM reverse primer
- 3 μl nanopure water
- 5 μl Taq (we used EconoTaq Green Taq)
- Ran for 30 cycles of denaturing, annealing, extension
- Initial denature @ 94C for 2 min.
- Denature @ 94C for 30 sec.
- Anneal @ 54, 58, 62C for 30 sec.
- Extend @ 72C for 90 sec.
- Final extension @ 72C for 10 min.
- Held @ 4C inifinitly.
(Note: same primers as before)
Restriction digest
- Sequential digest of pRL1383a and ccdB with HindIII then BamHI in NEBuffer 2 + BSA
Subcloning of lac device and BioBrick MCS in BB-pRL1383a
Restriction digest
- Sequential digest of pRL1383a and J33207/MCS segment with BamHI then HindIII
- Gel purified
Ligation
Results
Transformation into DH5α
Sixty colonies were observed for GFP fusion and there were too many colonies to count (>300) for the modified pRL1383a.
Verification of transformants
A colony PCR was conducted to verify the presence of the GFP fusion brick containing plasmid and the modified pRL1383a. PCR products observed on the gel were of the expected size. One band was observed for pRL1383a (MCS=~50bp + ~250bp(upstream/downstream verification regions)) and the GFP fusion brick (720bp + ~250bp(upstream/downstream verification regions).
Sequencing
Sequencing by the Greenwood Molecular Biology Facility returned the following sequences:
GFP fusion
pRL1383a multiple cloning site
Sequencing returned results that did not match the expected VF-VR from B0034. When Blasted against the parts registry as well as the NCBI database, no significant results were found. Currently, we have no idea what the sequence is. The read was dirty so perhaps the results were just that of a bad read sample?
Attempt #2
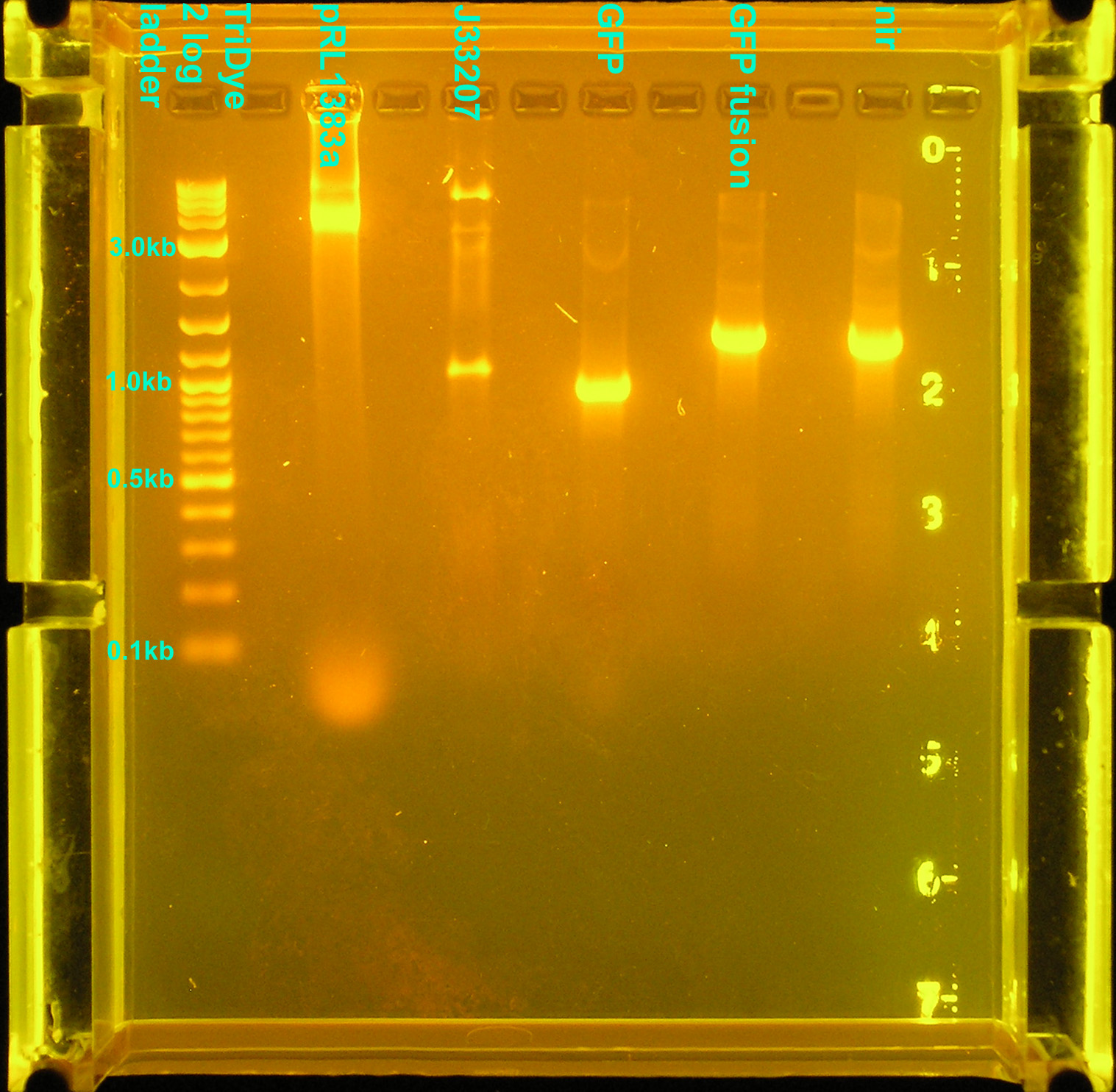
Restriction digests
Smear between 8-10kb may be due to too much pRL1383a plasmid? Two bands were observed for the RE digest of the J33207 PCR product. One band, ~1kb, is the size of the expected DNA fragment. The larger band, ~9kb, is yet unidentified.
Discussion
Success! GFP fusion has been added to BioBrick registry and will be submitted at a later date. Need to redo pRL1383a assembly.
ccdB has a BamHI site within the gene. Oops! Switch to J33207 (lac promoter + rbs + lacZ) for screening of successfully ligated plasmid in transformants.
 "
"