Team:Hawaii/Plasmid Prep
From 2008.igem.org
(→Second attempt on 2008.06.19) |
Normanwang (Talk | contribs) (→Results) |
||
| Line 27: | Line 27: | ||
== Results == | == Results == | ||
=== First attempt on 2008.06.17 === | === First attempt on 2008.06.17 === | ||
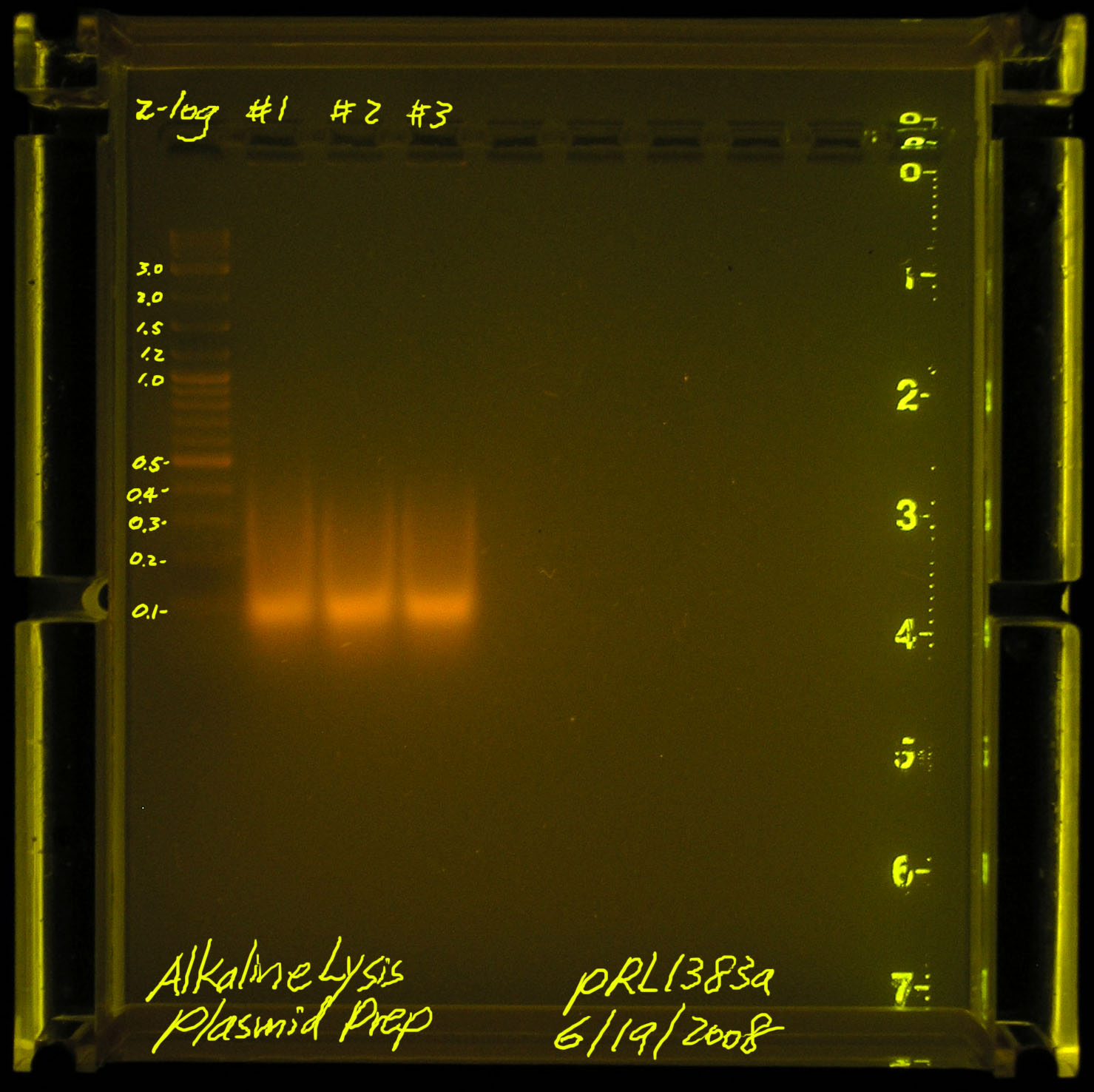
| - | [[Image:20080619-alkaline_lysis_plasmid_prep.jpg|thumb|left|Failed, we should see something bigger than a smear between 100kb-500kb]] | + | [[Image:20080619-alkaline_lysis_plasmid_prep.jpg|thumb|left|Failed, we should see something bigger than a smear between 100kb-500kb, this run uses 10ul plasmid-prep, 2ul loading dye]] |
| - | According to Gernot, we may have sheared the plasmid DNA (he has not seen the picture). Another problem may be that pRL1383a is a LOW copy plasmid. we should start with more material instead of the standard 1.5mL spindown per 30ul of TE suspended final plasmid. | + | According to Gernot, we may have sheared the plasmid DNA (he has not seen the picture). Another problem may be that pRL1383a is a LOW copy plasmid. we should start with more material instead of the standard 1.5mL spindown per 30ul of TE suspended final plasmid.<br/> |
=== Second attempt on 2008.06.19 === | === Second attempt on 2008.06.19 === | ||
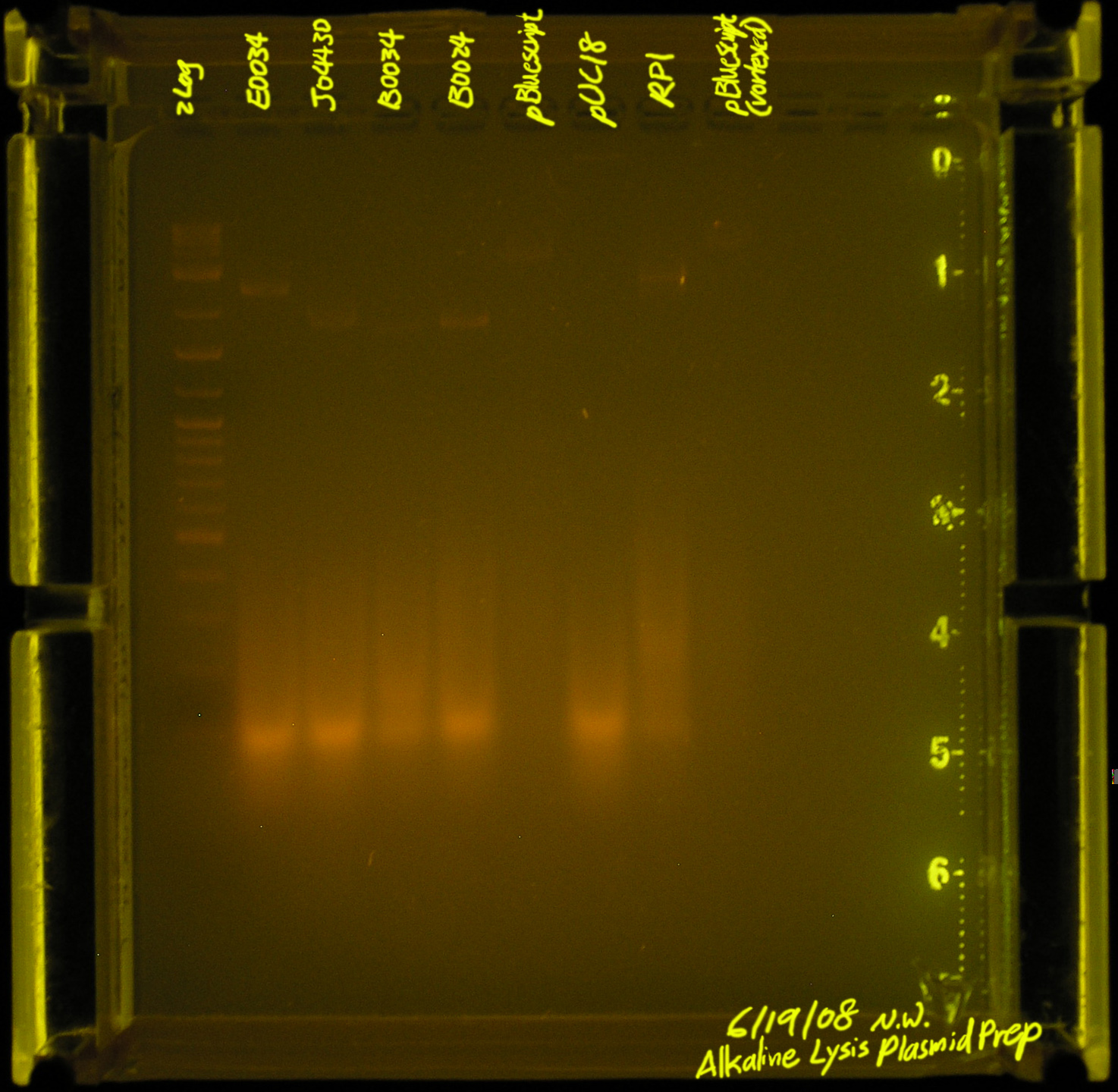
| - | + | [[Image:20080619-alkaline_lysis_plasmid_prep2.jpg|thumb|left|What is that stuff between 100bp and 500bp? this run is half as bright because i'm using 5ul plasmid-prep, 2ul loading dye]] | |
| - | + | ||
| - | + | ||
| - | + | ||
We prepped biobrick plasmids (high copy?) (assuming these clones have the parts we want on them): | We prepped biobrick plasmids (high copy?) (assuming these clones have the parts we want on them): | ||
* E0040 | * E0040 | ||
| Line 40: | Line 37: | ||
* B0034 | * B0034 | ||
* B0024 | * B0024 | ||
| - | + | Regular Plasmids | |
| - | + | * pBluescript | |
| + | * pUC18 | ||
| + | * RP1 | ||
| + | * pBluescript (vortexed, pippetted up down few times when loading, to see if it's the cause of the small fragments.) | ||
== Discusstion == | == Discusstion == | ||
Revision as of 09:43, 20 June 2008
Contents |
Protocol
1. Grow single colony of E. coli at 37C overnight in 5 ml LB w/ antibiotic selection.
2. Microcentrifuge 1.5 ml cells for 20 sec at 16,000g. Discard supernatant.
3. Resuspend pellet in 100 μl GTE solution.
- 50 mM glucose
- 10 mM EDTA
- 25 mM Tris-HCl (pH 8.0)
4. Let sit for 5 min. at room temperature.
5. Add 200 μl NaOH/SDS solution.
- 0.2 M NaOH
- 1% SDS
6. Mix by tapping tube.
7. Incubate on ice for 5 min.
8. Add 150 μl potassium acetate solution.
9. Invert a few times to mix.
10. Incubate on ice for 5 min.
11. Microcentrifuge for 3 min. at 16,000g.
12. Transfer supernatant to a new tube.
13. Add 0.8 ml 95% ethanol.
14. Incubate for 2 min. at room temperature.
15. Microcentrifuge for 1 min. at 16,000g at room temperature. Remove supernatant.
16. Wash pellet w/ 1 ml 70% ethanol. Aspirate to dry (dry in hood).
17. Resuspend pellet in 30 μl TE buffer.
Reference: Short Protocols in Molecular Biology
Results
First attempt on 2008.06.17
According to Gernot, we may have sheared the plasmid DNA (he has not seen the picture). Another problem may be that pRL1383a is a LOW copy plasmid. we should start with more material instead of the standard 1.5mL spindown per 30ul of TE suspended final plasmid.
Second attempt on 2008.06.19
We prepped biobrick plasmids (high copy?) (assuming these clones have the parts we want on them):
- E0040
- J04430
- B0034
- B0024
Regular Plasmids
- pBluescript
- pUC18
- RP1
- pBluescript (vortexed, pippetted up down few times when loading, to see if it's the cause of the small fragments.)
 "
"