Team:Heidelberg/Notebook/Killing I/Notebook/week4
From 2008.igem.org


| << Week 3 | Overview | Week 5 >> |
|---|
Week 4
Contents |
Monday, 08/25/08
project planning
- we finally received the strains sent to us by Prof. Bagdasarian. Beginning of next week we should also receive those from Prof. Derbyshire.
- this means we have to do mini(or maxi)-preps to get the plasmids from which we can amplify the oriT and the helper plasmids (RP1/4, pUB307 if we need this).
- We received the strains from Prof. Derbyshire. He sent us pUB307, RSF1010, pED350, pED361, pED369 and pED374.
- we received two plates with bacteria containing pUB307. We picked 4 colonies and inoculated them in with canamycin.
- we received the glycerol stocks of the cI from the registry and inoculated these for a maxiprep in LB with ampicilin.
Phage cloning strategy one
- Digestion of the small XbaI-XhoI lambda-fragment with AgeI
18µl DNA (from gel purification kit) 5µl AgeI 5µl NEB4 4µl water
- the digestion of the small lambda fragment showed no DNA on the gel...
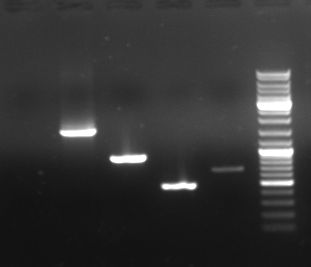
- we amplified GAM, GFP, Chloramphenicol and the OriT from Prof. Bagdasarian via PCR:
2µl Forward Primer 2µl Reverse Primer 25µl Phusion Mastermix 10ng DNA Template fill up to 50µl with water
PCR protocol 98°C 1min 98°C 30s | 55°C 30s | 25x 72°C 90s | 72°C 5min
- Results of PCR:
- lane2: GAM fragment (~1.44kb)
- lane3: GFP fragment (~0.92kb)
- lane4: oriT fragment (~0.52kb)
- lane5: CmR fragment (~0.66kb)
- lane6: DNA ladder mix
- NOTE: we have to find suitable names for all our OriT-Sequences and Helper Plasmids from the different research groups!!!
- Retransformation of the plasmids received from Prof. Derbyshire (he recommended to do this, because the DNA is very old)
- pUB307 - Canamycin resistance
- pED369 - Ampicilin resistance
- pED350 - Amp
- pEd361 - Amp
- pED374 - Amp
- We need streptomycin or sulphonamides to select the RSF1010...
- Digestion of pUB307 (Lanka) (4 minipreps), J01101 (cI) and small fragment (XbaI/XhoI)
- lane0: DNA ladder mix
- lane1: pUB307 (1) undigested
- lane2: pUB307 (1) EcoRI
- lane3: pUB307 (2) undigested
- lane4: pUB307 (2) EcoRI
- lane5: pUB307 (3) EcoRI
- lane6: pUB307 (4) EcoRI
- lane7: cI undigested (3kb)
- lane8: cI SfcI/HinDIII (1365/678/451/386/191)
- lane9: small fragment AgeI (1,7kb/7,3kb)
- Maxiprep
- cI
- pED 374
- Miniprep
- pED 361
- pED 350
- Nanodrop measurements
- CB2633 (OriT) 26,5 ng/µl; 2,25
- CB1133 (RP4) 39,6 ng/µl; 1,91
- CI Maxi 771,4 ng/µl, 1,93
- pUB207 ("Lanka")
- assay 1 50,8 ng/µl 2,02
- assay 1 60,5 ng/µl 2,02
- assay 1 62,7 ng/µl 1,97
- assay 1 62,7 ng/µl 2,02
- pED 361 42,7 ng/µl; 1,94
- pED 374 (Maxi) 87,7 ng/µl; 2,09
- pED 350 47,7 ng/µl; 2,13
- pUB 307 (Maxi) 269,4 ng/µl; 2,03
- pED 369 (Maxi) 383,5 ng/µl; 1,96
- Lambda cI 857 chemical transformation into DH5alpha and Top10 - no colony
- make DH5alpha and Top10 MgSO4-Stock
- 100ul competent bacteria + 4ml StandardI/0.2%Maltose
- 37C 3h
- 8min 3750rpm RT
- pellet dissolved in 3ml 0.01M MgSO4
- This Stock can be needed for phage infection, 10ul for one infection setting.
- This Stock can be used up to 1week (stored by RT)
- Lambda cI 857(Chris) amplification
- cut one plaque from the softgel, incubate in 0.5ml LambdaBuffer + 2 drops Chloroform, RT, 4.5h
- 50ul of the phage (see last line)+10ul DH5alpha MgSO4-Stock+200ul LambdaBuffer, 37C, 25min
- platte NZY softgel with agarose
NZY medium 10g/L NZ-amine (casein) 5g/L Yeastextract 5g/L NaCl 2g/L MgSO4 for gel: +1.5% agar/agarose for softgel: +0.7% agar/agarose + 0.2%maltose
- no plaque
- Lambda cI- ZMBH amplification
- 1ul phage LambdacI-ZMBH +10ul DH5alpha MgSO4-Stock+200ul LambdaBuffer, 37C, 25min
- platte NZY softgel with agarose
- completly lysed on the follow day
Tuesday, 08/26/08
Phage cloning strategy one
- Lambda cI 857 transformation
- chemical into MG1655, Top10
- electroporation into Top 10 F' of AG Mayer
- 5 plaques by E-Top10 (maybe), 1plaque by MG1655 (maybe)
- DNA purification of cI- ZMBH (see last day)
- follow kit, dissolve the DNA pellet in 200ul ddH2O + 2ul 1M Tris (8.0)
- we have a band!!!! concentration: maybe 1ng/µl
Wednesday, 08/27/08
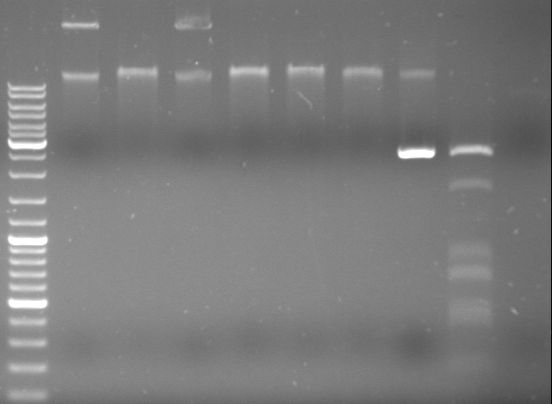
- repetition of PCR of GAM (2 vials), oriT (2), GFP (2) and CmR (3 vials)
- results:
- lane0: ladder
- lane1: GAM fragment (~1.44kb)
- lane2: GAM fragment (~1.44kb)
- lane3: oriT fragment (~0.52kb)
- lane4: oriT fragment (~0.52kb)
- lane5: GFP fragment (~0.92kb)
- lane6: GFP fragment (~0.92kb)
- lane7: CmR fragment (~0.66kb)
- lane8: CmR fragment (~0.66kb)
- lane9: CmR fragment (~0.66kb)
- lane10: blank
- lane11: DNA ladder mix
- no band in oriT and GFP visible --> reload these samples on another gel
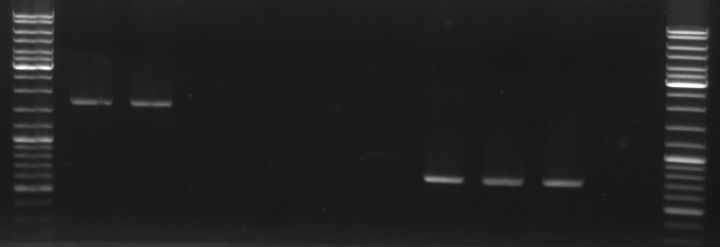
- lane0: DNA ladder mix
- lane1: oriT fragment (~0.52kb)
- lane2: oriT fragment (~0.52kb)
- lane3: GFP fragment (~0.92kb)
- lane4: GFP fragment (~0.92kb)
- Maxiprep
- pUB 307
- pED 369
- cI sent to GATC for sequenzing
- digestion of lambda-DNA with XhoI-XbaI following the old protocol
- if AgeI still doesn't work, we're going to have to check if we got the restriction site right and/or test the enzyme with another plasmid
- Gel
- lane2: ladder
- lane3: digestion of lambda-DNA with XhoI-XbaI
- lane 4 - 8: assays of Yin
- the gel was somehow damaged when we took out of the gel chamber. it was impossible to go on working from it
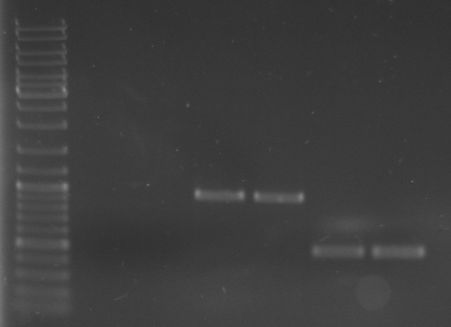
- Gel
- lane 0: DNA ladder mix
- lane 1: pED374 1 (Purified PCR Product)
- lane 2: pED374 2 (Purified PCR Product)
- lane 3: GFP 1 (Purified PCR Product)
- lane 4: GFP 2 (Purified PCR Product)
- lane 5: OriT 1 (Purified PCR Product)
- lane 6: OriT 2 (Purified PCR Product)
- Overnight digestion
- Lambda cI 857 with XbaI-XhoI
- lambda ZMBH cI- digestion with XbaI-XhoI
- lambda DNA with AgeI to test the enzyme
- Lambda cI- ZMBH digestion with XbaI and XhoI
- no fragment. Lambda cI- ZMBH is a bit smaller than Lambda cI 857
- Lambda cI 857 Amplification
- plaques by electroporation (see the day before)
- one plaque by chemical transformation MG1655
- cut and incubate the plaque (see Monday)by 4°C overnight
Thursday, 08/28/08
cI
- the sequence of the cI we received from MIT was wrong and corresponded to GFP instead. We will do another prep this evening so that we can send the "cI" to GATC again tomorrow.
Phage cloning strategy one
- Glycerolstocks of pBAD33 and pBluescript
- in the freezer in the cellar
- 1ml bacteria culture, 150 µl Glycerol 80%, vortex, let stand for one hour
- Maxiprep
- pBAD33
- pBluescript
- Miniprep
- cI
- infection of dh5alpha by Lambda cI 857 (supernatant of the plaque incubate, see the day before)
- one plate with eletroporation-Lambda cI 857, one plate with chemical transformation Mg1655-Lambda cI 857
- complettly lysed on the follow day
Friday, 08/29/08
cI
- cI sent to GATC again)
Phage cloning strategy one
- Lambda DNA purification (follow kit, like Tue)
- run the DNA in 1% gel: no signal
- no DNA? to low concentration? lost the DNA by the purification?
- In vitro Packaging
- follow kit, pack Lambda cI 857
- titering with DH5alpha, MG1655
- DH5alpha: very nice infection, a lot of plaques. titer:10^7puf/ml
- MG1655: the infectionrate is lower than with DH5alpha, but also many plaques. Titer: 4*10^6pmf/ml
Teammeeting in the afternoon
| << Week 3 | Overview | Week 5 >> |
|---|
 "
"