Team:Imperial College/Fluorescence
From 2008.igem.org
(Difference between revisions)
m |
|||
| Line 38: | Line 38: | ||
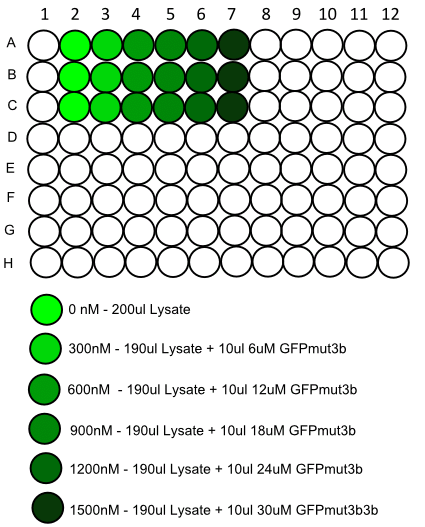
#Repeat this dilution three times to enable us to average out any errors. | #Repeat this dilution three times to enable us to average out any errors. | ||
#Once the lysis is complete we are ready to carry out the dilutions of GFPmut3b in lysed ''B. subtilis'', | #Once the lysis is complete we are ready to carry out the dilutions of GFPmut3b in lysed ''B. subtilis'', | ||
| - | #Then follow the plate loading schematic below being careful to avoid bubbles and mix solutions thoroughly before adding to the plate | + | #Then follow the plate loading schematic below being careful to avoid bubbles and mix solutions thoroughly before adding to the plate. |
#Place in the plate reader and load up as follows: | #Place in the plate reader and load up as follows: | ||
| Line 45: | Line 45: | ||
|}} | |}} | ||
| - | {{Imperial/EndPage|Protocols|}} | + | {{Imperial/EndPage|Protocols|Protocols}} |
Latest revision as of 17:42, 28 October 2008
|
|||||||
 "
"