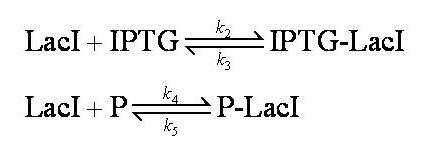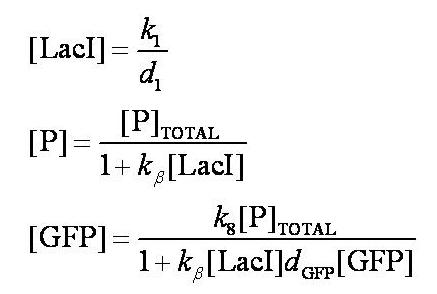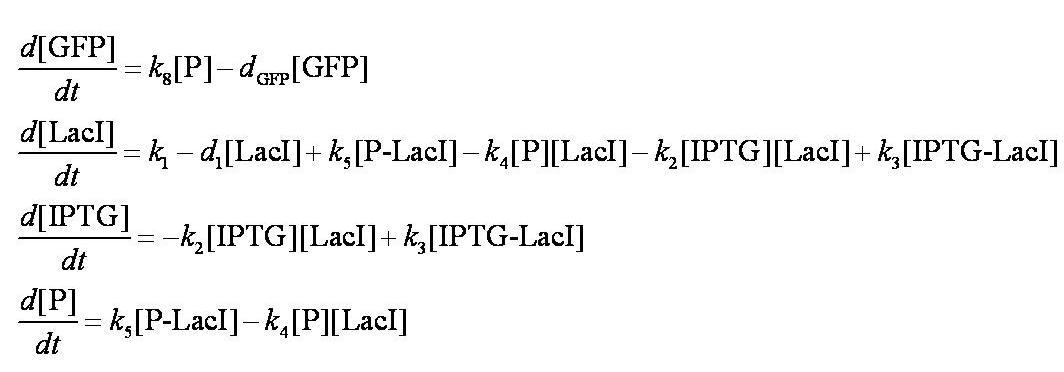Team:Imperial College/Genetic Circuit Details
From 2008.igem.org
| Line 27: | Line 27: | ||
These are evaluated numerically using Matlab's ODE solver. | These are evaluated numerically using Matlab's ODE solver. | ||
| - | Note from the differential equations above that the steady-state concentration of free promoter will be independent of the concentration of IPTG introduced into the system. The pre-steady-state maximum concentrotion of GFP attained will differ, however - this is illustrated on the [[Team:Imperial_College/Genetic_Circuit | Genetic Circuit]] page and can be explored further using the | + | Note from the differential equations above that the steady-state concentration of free promoter will be independent of the concentration of IPTG introduced into the system. The pre-steady-state maximum concentrotion of GFP attained will differ, however - this is illustrated on the [[Team:Imperial_College/Genetic_Circuit | Genetic Circuit]] page and can be explored further using the [[Media:InduciblePromoterSimple.m|simulations]]. |
}} | }} | ||
Revision as of 00:22, 29 October 2008
|
|||||||||||||||
 "
"