Team:Imperial College/Major Results
From 2008.igem.org
| Line 17: | Line 17: | ||
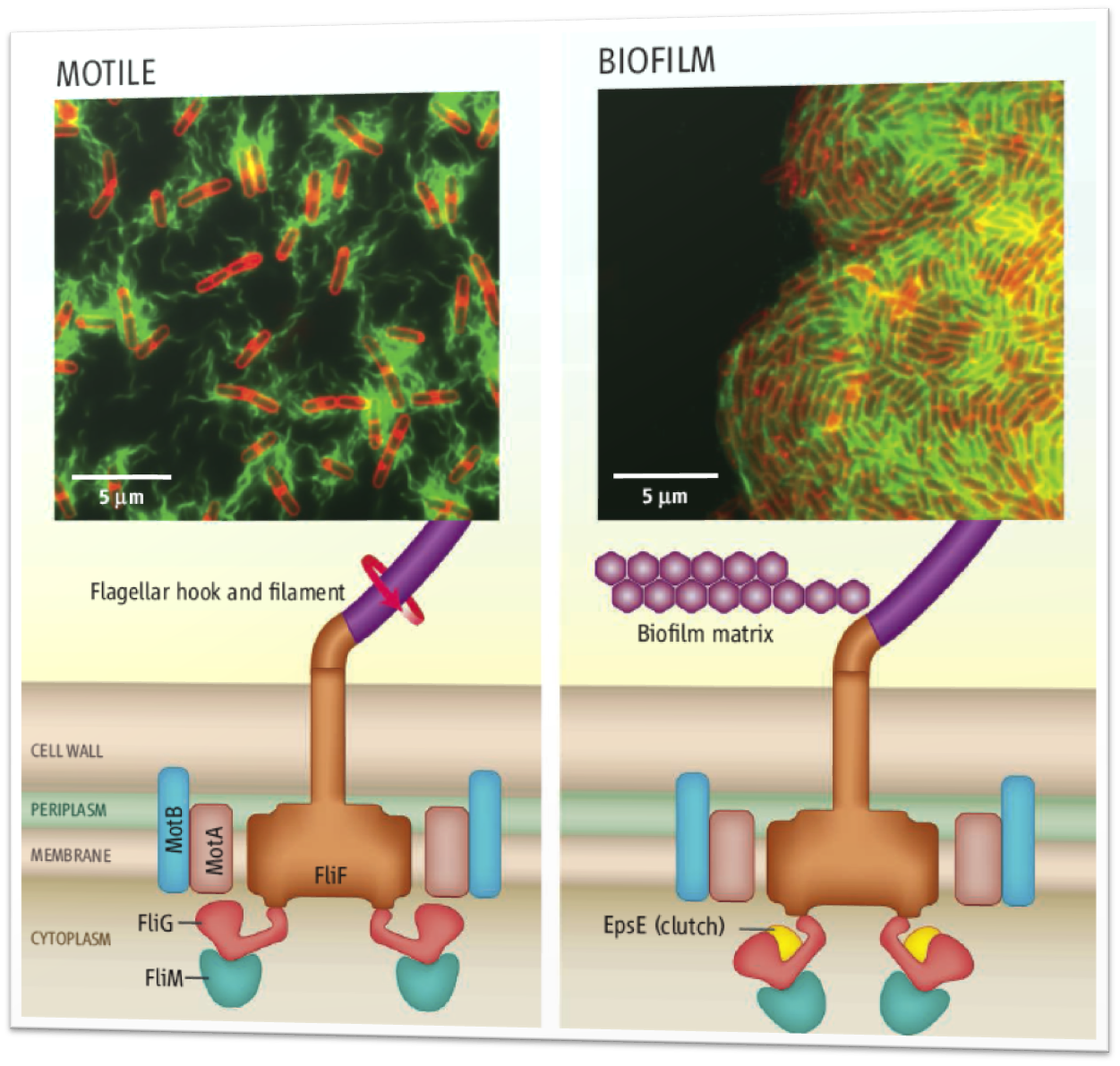
We studied the motility of ''B. subtilis'' and developed an elegant mechanical model to describe cell trajectory. We fitted our model to our laboratory data and discovered that our model matches cell trajectory data extremely well. | We studied the motility of ''B. subtilis'' and developed an elegant mechanical model to describe cell trajectory. We fitted our model to our laboratory data and discovered that our model matches cell trajectory data extremely well. | ||
<br><br> | <br><br> | ||
| - | <center>[[Image:GFP2.gif]]</center | + | <center>[[Image:GFP2.gif]]</center><br>}} |
{{Imperial/Box1|Characterisation of Chloramphenicol Acetyltransferase|<br> | {{Imperial/Box1|Characterisation of Chloramphenicol Acetyltransferase|<br> | ||
Revision as of 22:18, 29 October 2008
Experimental Results
|
|||||||||||||||||||||||||||
 "
"