Team:Imperial College/Motility
From 2008.igem.org
(Difference between revisions)
m |
m |
||
| Line 4: | Line 4: | ||
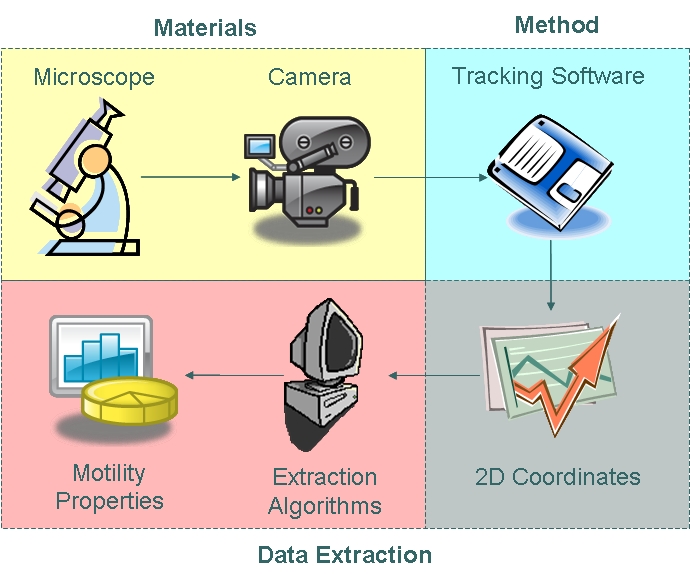
{{Imperial/Box2|Motility Analysis|[[Image:Approach.jpg|450px|center]] | {{Imperial/Box2|Motility Analysis|[[Image:Approach.jpg|450px|center]] | ||
<br><br> | <br><br> | ||
| - | The motility of ''B. | + | The motility of ''B. subtilis'' is hypothesised to be affected by various levels of EpsE expression. In order to model motility as a function of EpsE production, we have decided to use video microscopy techniques to analyse the motility of ''B. subtilis''. We hope to obtain a transfer function model relating EpsE expression to bacterial motility characteristics such as run velocity, run duration, tumbling angle and tumbling duration. This modelling process is shown on the right and has been divided into 3 main sections: |
#'''[[/Validation | Validation of Tracking Software ]]''' | #'''[[/Validation | Validation of Tracking Software ]]''' | ||
#*In order to assess the error associated with tracking algorithms applied to a digitised images sequence, a series of steps were taken to '''[[IGEM:IMPERIAL/2008/Prototype/Drylab/Validation |validate]]''' the tracking software. | #*In order to assess the error associated with tracking algorithms applied to a digitised images sequence, a series of steps were taken to '''[[IGEM:IMPERIAL/2008/Prototype/Drylab/Validation |validate]]''' the tracking software. | ||
#'''[[/Motility_data_collection|Motility Data Acquisition]]''' | #'''[[/Motility_data_collection|Motility Data Acquisition]]''' | ||
| - | #*Data on run velocity, run duration, tumbling angle and | + | #*Data on run velocity, run duration, tumbling angle and tumbling duration were extracted from coordinate data output provided by the tracking software as part of the process of gathering data. |
#'''[[/Model_Fitting|Model Fitting]]''' | #'''[[/Model_Fitting|Model Fitting]]''' | ||
#*Several alternative models were created for the purpose of model fitting. The motility data obtained is then analysed and fitted to alternative models. Preferences will then be assigned to fitted models using probabilistic methods such as Bayesian Analysis. | #*Several alternative models were created for the purpose of model fitting. The motility data obtained is then analysed and fitted to alternative models. Preferences will then be assigned to fitted models using probabilistic methods such as Bayesian Analysis. | ||
Revision as of 16:45, 15 October 2008
|
|||||||
 "
"