Team:Imperial College/Summary
From 2008.igem.org
m |
|||
| (22 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Imperial/StartPage2}} | {{Imperial/StartPage2}} | ||
| - | === | + | |
| + | === Project Summary === | ||
{{Imperial/Box1|Design| | {{Imperial/Box1|Design| | ||
In order to achieve our specifications of design, we require the following devices; | In order to achieve our specifications of design, we require the following devices; | ||
| Line 13: | Line 14: | ||
(AB is our antibiotic resistance cassette, ''ytvA'' is the gene controlling the light-sensing pathway, ''SB'' is the biomaterial, ''epsE'' the clutch and the 5' and 3' sections are integration sites. Light-inducible promoters are labelled with an 'L') | (AB is our antibiotic resistance cassette, ''ytvA'' is the gene controlling the light-sensing pathway, ''SB'' is the biomaterial, ''epsE'' the clutch and the 5' and 3' sections are integration sites. Light-inducible promoters are labelled with an 'L') | ||
| - | |||
| - | |||
|}} | |}} | ||
| - | {{Imperial/Box1| | + | {{Imperial/Box1|Modelling| |
| + | The Dry Lab used computational simulations to explore the different properties of the Biofabricator. Our activities are summarized on this page. To find out more please visit the [https://2008.igem.org/Team:Imperial_College/Dry_Lab '''Dry Lab Hub''']. | ||
====== Growth Curve ====== | ====== Growth Curve ====== | ||
| - | + | We have developed a simple model for the growth of B. subtilis where the rate of growth is related to the amount of nutrients available. To this purpose we have exploited the ideas put forward by last year's Imperial College iGEM team for their modelling of F2620 in a cell-free system. | |
====== Genetic Circuit ====== | ====== Genetic Circuit ====== | ||
| - | We have | + | We have also built mathematical models for the time evolution of the basic genetic circuits that comprise our device. We have verified which model best describe the behaviour of the circuit better by using laboratory data. |
| - | + | ||
====== Motility Analysis ====== | ====== Motility Analysis ====== | ||
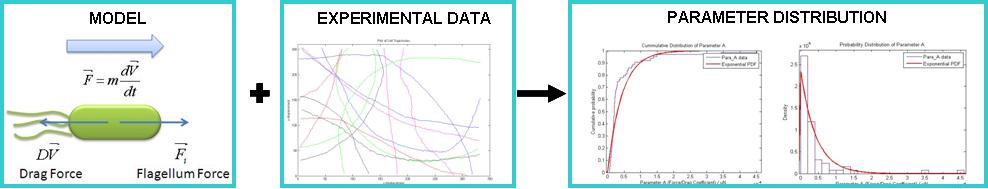
| - | + | Finally, we have carried out a detailed analysis of the swimming motility of B. subtilis, which led us, among other things, to develop a simple mechanical model for the swimming motility of B. subtilis. Using manual tracking, we were able to extract x,y coordinate data from the cell trajectory. This has allowed us to fit experimental data with our model. The data suggest that flagellar force of ''B. subtilis'' is Exponentially distributed. | |
| + | |||
| + | All model simulations and motility data analysis were carried out with MATLAB. Cell tracking was done with ImageJ via the Manual Tracking Plugin. All our MATLAB files can be found in the Appendices section. | ||
| + | |||
[[Image:Motility_Summary.jpg|center|600px]]<br> | [[Image:Motility_Summary.jpg|center|600px]]<br> | ||
|}} | |}} | ||
| Line 32: | Line 34: | ||
[[Image:Implementation.PNG|center|600px]] | [[Image:Implementation.PNG|center|600px]] | ||
| - | }} | + | |}} |
{{Imperial/Box1|Testing| | {{Imperial/Box1|Testing| | ||
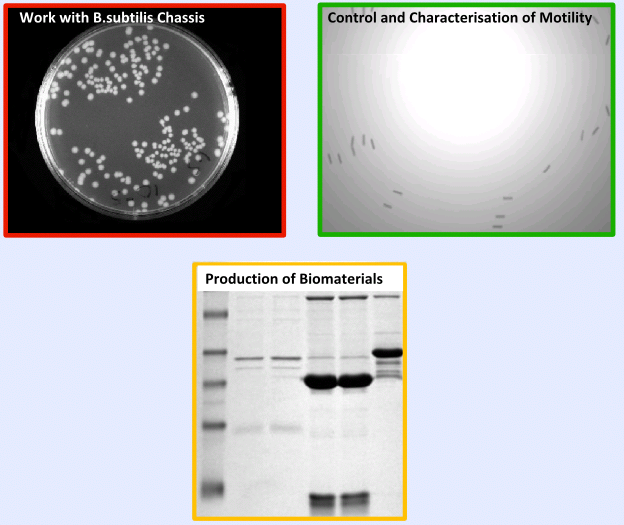
The testing and validation of our project can be split into three main areas; | The testing and validation of our project can be split into three main areas; | ||
*'''Work with ''B. subtilis''''' - Including characterisation of growth curves and transformation, | *'''Work with ''B. subtilis''''' - Including characterisation of growth curves and transformation, | ||
| - | *'''Characterisation | + | *'''Extensive Characterisation ''' of new ''B.subtilis'' biobricks, Chloramphenicol resistance gene and motility |
*'''Production of Biomaterials in ''B. subtilis''''' | *'''Production of Biomaterials in ''B. subtilis''''' | ||
| - | + | If you'd like to see more information on the key results from the testing and validation, you can find it on the [https://2008.igem.org/Team:Imperial_College/Major_Results '''Results Page''']. | |
| - | + | =====Results===== | |
| - | |[[Image:Result.PNG|center|300px]] | + | |[[Image:Result.PNG|center|300px]]}} |
| - | }} | + | |
{{Imperial/Box1|Achievements| | {{Imperial/Box1|Achievements| | ||
Here is a summary of the achievements of the Imperial College 2008 team: | Here is a summary of the achievements of the Imperial College 2008 team: | ||
| - | *Submitted | + | *Submitted 45 documented parts to the Registry |
| - | *Characterized and improved the existing part..... | + | *<html><a target="_blank" href="https://2008.igem.org/Team:Imperial_College/CAT"><b>Characterized</b></a></html> and improved the existing part <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_J31005" target="_blank">BBa J31005</a></html> (chloramphenicol acetyl transferase, CAT) |
| + | *<html><a target="_blank" href="https://2008.igem.org/Team:Imperial_College/Biobricks"><b>Characterized</b></a></html> the new promoter and ribosome binding sites biobricks <html><a target="_blank" href="http://partsregistry.org/wiki/index.php/Part:BBa_K143079">BBa K143079</a></html> and <html><a target="_blank" href="http://partsregistry.org/wiki/index.php/Part:BBa_K143082">BBa K143082</a></html> that we submitted this year. | ||
*Developed integration sequences for Biobricks, to allow devices to be constructed that can then be excised and planted into ''B. subtilis'' | *Developed integration sequences for Biobricks, to allow devices to be constructed that can then be excised and planted into ''B. subtilis'' | ||
| - | * | + | *Laid the groundwork for future teams to work with ''B. subtilis'' by BioBricking and characterising promoters, RBSs, integration sequences, coding sequences and complex devices |
*Showed that expansion into other organisms is a definite possibility! | *Showed that expansion into other organisms is a definite possibility! | ||
*Developed a method for tracking and analysing bacterial motility | *Developed a method for tracking and analysing bacterial motility | ||
*Helped Bristol by sending them a mini-iGEM project: ''Chemotactic dot-to-dot'' with information on quorum sensing and directed movement | *Helped Bristol by sending them a mini-iGEM project: ''Chemotactic dot-to-dot'' with information on quorum sensing and directed movement | ||
| - | *Helped Bristol by sending them | + | *Helped Bristol by sending them part <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_J37015" target="_blank">BBa_J37015</a></html> (AHL generator + GFP) from our 2006 stock which was an empty vector in the Registry |
| - | }} | + | *Helped Cambridge by sending them a plate of ''Synechocystis'' PCC680 and a genome preparation |
| + | |}} | ||
<hr><br> | <hr><br> | ||
| - | {{Imperial/Box2||Of course, that is a very simplified description of our project. We expanded upon our project by looking into possible areas for real-world applications. For a case-study of such an implementation, check out how our project fits in with [[Team:Imperial_College/Cellulose | '''>>> Biocouture >>>''']]}} | + | {{Imperial/Box2||Of course, that is a very simplified description of our project. We expanded upon our project by looking into possible areas for real-world applications. For a case-study of such an implementation, check out how our project fits in with [[Team:Imperial_College/Cellulose | '''>>> Biocouture >>>''']]|}} |
{{Imperial/EndPage|Chassis_2|Cellulose}} | {{Imperial/EndPage|Chassis_2|Cellulose}} | ||
Latest revision as of 03:12, 30 October 2008
Project Summary
|
|||||||||||||||||||||||||||
 "
"