Team:Imperial College/Summary
From 2008.igem.org
m |
m |
||
| Line 2: | Line 2: | ||
=== Summer Summary === | === Summer Summary === | ||
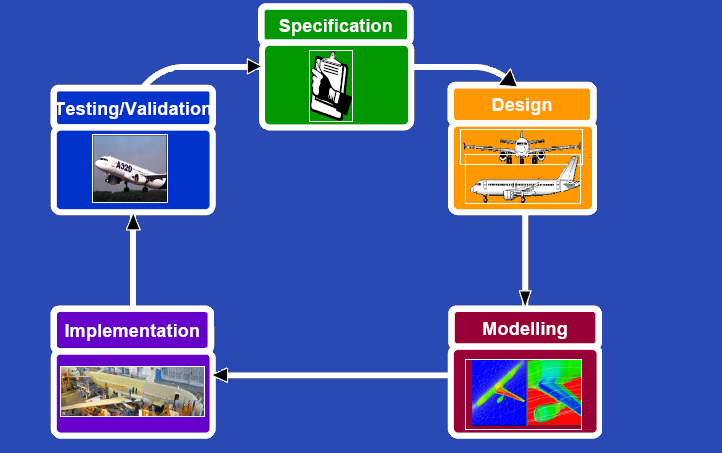
| - | {{Imperial/Box2|| | + | {{Imperial/Box2|Our Approach|[[Image:Cycle.PNG|center|500px]]|}} |
| - | + | ||
| - | + | {{Imperial/Box1|Design| | |
| - | + | In order to achieve our specifications of design previously described, we require the following constructs; | |
| - | + | *'''Light sensing device''' - Converting a light input into an output of PoPS, | |
| - | + | *'''Biomaterial production device'''- Converting a PoPS input into an output of biomaterial, | |
| - | + | *'''Motility Control device''' - Converting a PoPS input into an output of motility arrest, | |
| - | + | *'''Integration device''' - To allow integration and selection of our genetic constructs, | |
| + | <br> | ||
Our basic approach was to constitutively upregulate a light-sensitive pathway in ''B. subtilis'', then use a downstream product from that pathway to selectively regulate expression of our clutch and biomaterial. This overview is shown on the right. To do this we need to produce at least one constitutive promoter to precede our light-sensing mechanism, as well as a 'light-inducible' promoter in front of our clutch and biomaterial. Additionally, we'll need to make all the other standard parts such as ribosome binding sites (RBS) and terminators. | Our basic approach was to constitutively upregulate a light-sensitive pathway in ''B. subtilis'', then use a downstream product from that pathway to selectively regulate expression of our clutch and biomaterial. This overview is shown on the right. To do this we need to produce at least one constitutive promoter to precede our light-sensing mechanism, as well as a 'light-inducible' promoter in front of our clutch and biomaterial. Additionally, we'll need to make all the other standard parts such as ribosome binding sites (RBS) and terminators. | ||
| - | + | [[Image:Genetic circuit.PNG|750px|center]] | |
| - | + | Here, AB is our antibiotic resistance cassette, ''ytvA'' is the gene controlling the light-sensing pathway, ''SB'' is the biomaterial, ''epsE'' the clutch and the 5' and 3' sections are integration sites. Light-inducible promoters are labelled with an 'L'. | |
| - | |||
|}} | |}} | ||
| - | {{Imperial/Box1| | + | {{Imperial/Box1|Modeling - Overview| |
Basically our dry lab team concentrated on characterising the chassis. In the dry lab section you'll find pages on the genetic circuit, growth curve and motility analysis from our project; this section will give a brief brief overview of each area (no pictures?). It will also include (in the motility analysis part) a movie (y/n?) of the motility response of ''B. subtilis'' with an inducible ''epsE'' gene BioBricked in. | Basically our dry lab team concentrated on characterising the chassis. In the dry lab section you'll find pages on the genetic circuit, growth curve and motility analysis from our project; this section will give a brief brief overview of each area (no pictures?). It will also include (in the motility analysis part) a movie (y/n?) of the motility response of ''B. subtilis'' with an inducible ''epsE'' gene BioBricked in. | ||
====== Growth Curve ====== | ====== Growth Curve ====== | ||
| Line 27: | Line 26: | ||
We feel accurate modelling of the genetic circuit contributes greatly to the characterisation of synthetic systems. As part of the project, the behaviours of constitutive and inducible promoters were modelled for comparison with our experimental data. | We feel accurate modelling of the genetic circuit contributes greatly to the characterisation of synthetic systems. As part of the project, the behaviours of constitutive and inducible promoters were modelled for comparison with our experimental data. | ||
====== Motility Analysis ====== | ====== Motility Analysis ====== | ||
| - | A major component of the system is the motility and shift between a motile and arrested state with the expression of EpsE | + | A major component of the system is the motility and shift between a motile and arrested state with the expression of EpsE. |
|}} | |}} | ||
| - | {{Imperial/Box1| | + | {{Imperial/Box1|Implementation| |
| + | Think what we can refer to as implementation is the construction of the parts. Last week Kirsten had the idea that we could make a big gel with the digests from all our parts, I will look into this week to get an image. \We could use this then put some stuff about no. of parts etc.}} | ||
| + | |||
| + | {{Imperial/Box1|Testing| | ||
The wet lab team was responsible for designing the constructs and setting up the cloning strategy to get us from the starting parts to the finished system. We were also responsible for designing and BioBricking the starting parts, of course, and designing and implementing the integration brick technique. This section shows some of the major results that came from the wet lab over the summer. Motility results can go in the dry lab overview above. | The wet lab team was responsible for designing the constructs and setting up the cloning strategy to get us from the starting parts to the finished system. We were also responsible for designing and BioBricking the starting parts, of course, and designing and implementing the integration brick technique. This section shows some of the major results that came from the wet lab over the summer. Motility results can go in the dry lab overview above. | ||
====== Transformation ====== | ====== Transformation ====== | ||
Revision as of 23:52, 20 October 2008
Summer Summary
|
|||||||||||||||||||||||||||||||
 "
"