Team:Imperial College/Summary
From 2008.igem.org
| Line 54: | Line 54: | ||
<hr><br> | <hr><br> | ||
{{Imperial/Box2||Of course, that is a very simplified description of our project. We expanded upon our project by looking into possible areas for real-world applications. | {{Imperial/Box2||Of course, that is a very simplified description of our project. We expanded upon our project by looking into possible areas for real-world applications. | ||
| + | <br> | ||
For a case-study of such an implementation, check out how our project fits in with [[Team:Imperial_College/Cellulose | '''>>> Biocouture >>>''']]}} | For a case-study of such an implementation, check out how our project fits in with [[Team:Imperial_College/Cellulose | '''>>> Biocouture >>>''']]}} | ||
{{Imperial/EndPage|Chassis_2|Cellulose}} | {{Imperial/EndPage|Chassis_2|Cellulose}} | ||
Revision as of 11:35, 26 October 2008
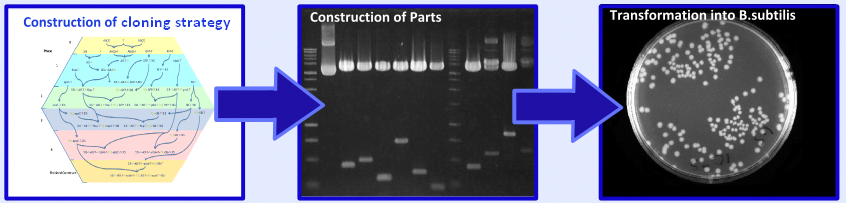
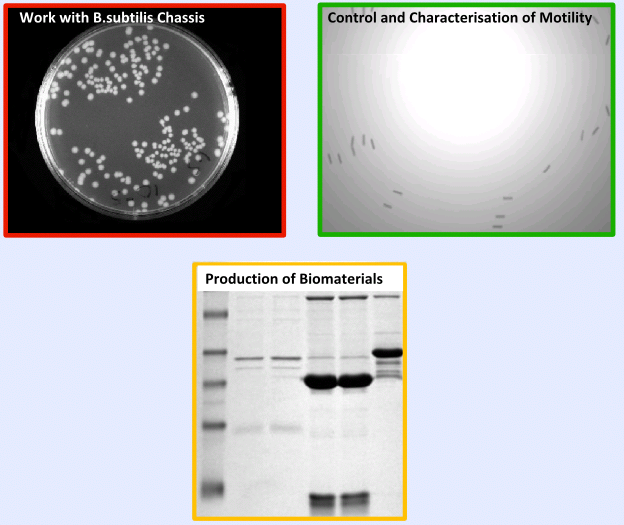
Summer Summary
|
|||||||||||||||||||||||||||
 "
"