Team:KULeuven/5 September 2008
From 2008.igem.org
(→Wet Lab) |
|||
| Line 1: | Line 1: | ||
{{:Team:KULeuven/Tools/Header}} | {{:Team:KULeuven/Tools/Header}} | ||
| + | |||
== Lab Work == | == Lab Work == | ||
| Line 5: | Line 6: | ||
=== Wet Lab === | === Wet Lab === | ||
| - | + | We had a very happy day in the lab today and this is what we did: | |
| + | * We put the digests of R0011+B0033, K145001+B0015, K145150 and K145013 on gel. They seemed OK and so we extracted them out of the gel. | ||
| + | * We made a Miniprep and digest of the following parts: <div style="margin-left:10px;"> cut with ''Eco''RI and ''Spe''I -> R0053+P0152, R0040+B0033, R0040+J23066, B0014+B0033 and R1052. </div> <div style="margin-left:10px;"> cut with XbaI -> R0040+E0240, C0061+B0015 and C0056+B0015. </div> | ||
| + | * We made a glycerolstock of the ligations that have suceeded so far (C0040+B0014, C0060+B0015, C0012+B0015, K145015+B0015, K145151+B0015 and R0040+E0240). | ||
| + | * We set up the following ligations: K145150+pSB1A2, K145150+E0240, K145150+B0034, K145013+pSB1A2 and (B0014+B0033)+(K145151+B0015). | ||
| + | * Unfortunately, there were also some failures. The PCR for T7 polymerase with UmuD tag failed again. The result of the PCR test of the transduction was also a bit dodgy and the digest of R1052 resulted in a smear (but apparently it is a crappy part). A final attempt to construct T7 polymerase with UmuD tag was started (PCR). | ||
| + | |||
| + | But the most exciting thing today, were our flourescent colonies! We constructed two OUTPUT-subsystems containing GFP with and without LVA tag, as can be seen in this picture. On the left side GFP with LVA tag (our part K145015): less fluorescent. On the right side GFP without LVA tag (part E0240): more fluorescent. This means that our output and GFP-LVA work - hurrah! | ||
[[Image:DSC02957.JPG|center|800px]] | [[Image:DSC02957.JPG|center|800px]] | ||
| - | Final Scheme | + | |
| + | |||
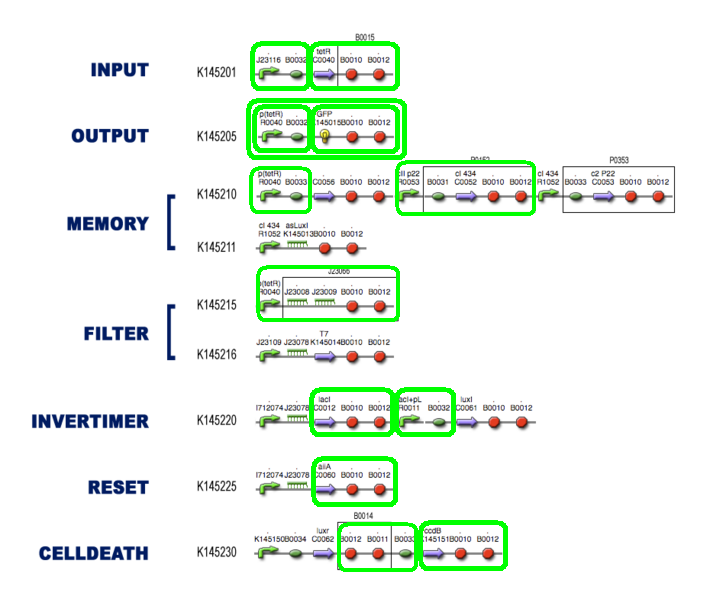
| + | Final Scheme so far: | ||
[[Image:5sept final.PNG|center|700px]] | [[Image:5sept final.PNG|center|700px]] | ||
| - | Parallel Scheme | + | Parallel Scheme so far: |
[[Image:5sept parallel.PNG|center|700px]] | [[Image:5sept parallel.PNG|center|700px]] | ||
| Line 19: | Line 29: | ||
NOT THAT MUCH MORE PARTS TO FINISH!!!! | NOT THAT MUCH MORE PARTS TO FINISH!!!! | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
=== Dry Lab === | === Dry Lab === | ||
Revision as of 20:04, 5 September 2008
Contents |
Lab Work
Wet Lab
We had a very happy day in the lab today and this is what we did:
- We put the digests of R0011+B0033, K145001+B0015, K145150 and K145013 on gel. They seemed OK and so we extracted them out of the gel.
- We made a Miniprep and digest of the following parts: cut with EcoRI and SpeI -> R0053+P0152, R0040+B0033, R0040+J23066, B0014+B0033 and R1052.cut with XbaI -> R0040+E0240, C0061+B0015 and C0056+B0015.
- We made a glycerolstock of the ligations that have suceeded so far (C0040+B0014, C0060+B0015, C0012+B0015, K145015+B0015, K145151+B0015 and R0040+E0240).
- We set up the following ligations: K145150+pSB1A2, K145150+E0240, K145150+B0034, K145013+pSB1A2 and (B0014+B0033)+(K145151+B0015).
- Unfortunately, there were also some failures. The PCR for T7 polymerase with UmuD tag failed again. The result of the PCR test of the transduction was also a bit dodgy and the digest of R1052 resulted in a smear (but apparently it is a crappy part). A final attempt to construct T7 polymerase with UmuD tag was started (PCR).
But the most exciting thing today, were our flourescent colonies! We constructed two OUTPUT-subsystems containing GFP with and without LVA tag, as can be seen in this picture. On the left side GFP with LVA tag (our part K145015): less fluorescent. On the right side GFP without LVA tag (part E0240): more fluorescent. This means that our output and GFP-LVA work - hurrah!
Final Scheme so far:
Parallel Scheme so far:
NOT THAT MUCH MORE PARTS TO FINISH!!!!
Dry Lab
Dr. Coli and his danger, ethics, ethics, ethics...
Modeling
SimBiology2Latex Toolbox has been finalized and can be found on the wiki.
MultiCell Toolbox has entered it's final design stages. A preview of the GUI can be found on the wiki.
Some more work on diffusion has been done. Sensitivity Analyses is still a pain in the ***.
Wiki
Homepage has been revamped, removing a lot of bugs. IE fixes still need to follow. Components bar has been fixed.
Remarks
Strip of the day
| << return to notebook | return to homepage >> | ||
| < previous friday | ← yesterday | tomorrow → | next monday > |
 "
"