Team:KULeuven/Concreties
From 2008.igem.org
(→Filter) |
(→Input) |
||
| Line 5: | Line 5: | ||
==Input== | ==Input== | ||
| + | |||
| + | The input could be abstracted by using an easily controllable light flash. This system uses the BioBricks of the Austin 2004 iGEM team. | ||
| + | |||
| + | |||
| + | For the use of that system, we need (see http://partsregistry.org/Featured_Parts:Light_Sensor): | ||
| + | * '''BBa_I15008''': makes biliverdin IXalpha out of the haem group | ||
| + | * '''BBa_I15009''': makes phycobilicyanin out of biliverdin IXalpha | ||
| + | These two parts are thus necessary to make phycobilicyanin, which is the chromophore needed for the proper working of the following part. | ||
| + | * '''BBa_I15010''': makes a chimeric Cph1 light receptor / EnvZ protein | ||
| + | The EnvZ kinase domain phosphorylates OmpR. Now the transcription is initiated by the fourth and last part: | ||
| + | * '''BBa_R0082''': this is a promoter that is activated by phosphorylated OmpR. | ||
| + | |||
| + | |||
| + | Notice that induction of this system could not be accomplished by a light flash, but should be done by a "''dark flash''". Indeed, red light (660 nm) inhibits phosphorylation of OmpR, and so inhibits the promoter. A dark flash results in OmpR phosphorylation, and so transcription from the OmpR-P dependent promoter. This is however not an ideal system, because keeping the E.coli in 660 nm light environment is not easy. | ||
| + | |||
| + | |||
| + | Notice also that as a result of this system, we need to work with EnvZ deficient E.coli. | ||
==Output== | ==Output== | ||
Revision as of 22:09, 2 July 2008
Op deze pagina verzamelen we concrete invullingen omtrent het project. De bedoeling is niet meer om over conceptuele dingen na te denken, maar om bijvoorbeeld op zoek te gaan naar analogen met vorige projecten, bruikbare BioBricks enzoverder. Voorlopig gooien we het nog niet onmiddellijk op de voorgrond, tot we meer voet in de aarde hebben omtrent haalbaarheid en zo.
Contents |
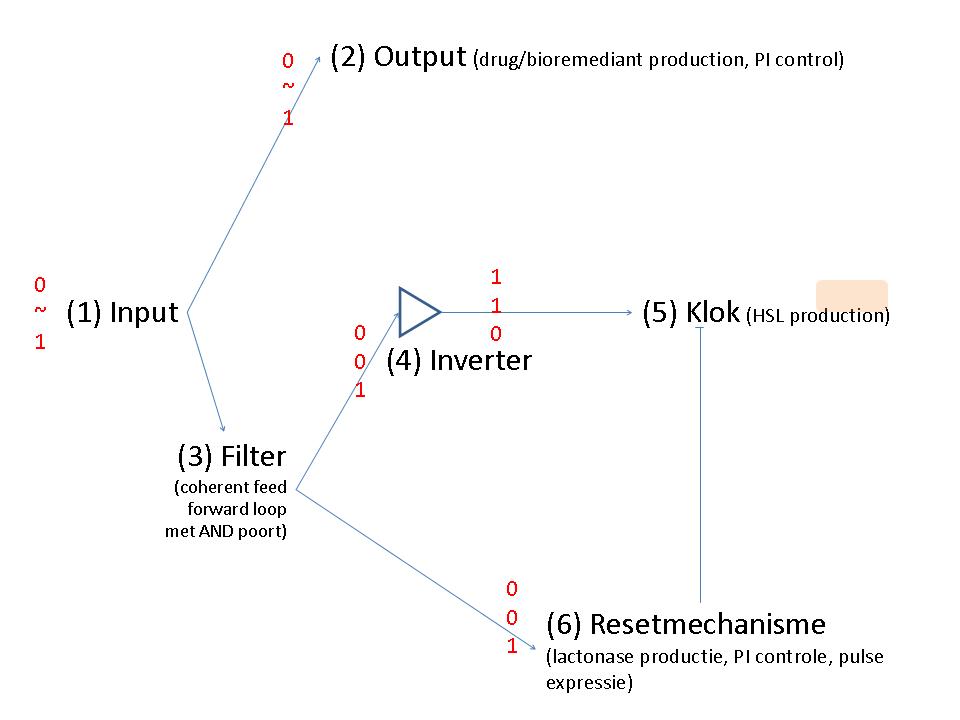
Input
The input could be abstracted by using an easily controllable light flash. This system uses the BioBricks of the Austin 2004 iGEM team.
For the use of that system, we need (see http://partsregistry.org/Featured_Parts:Light_Sensor):
- BBa_I15008: makes biliverdin IXalpha out of the haem group
- BBa_I15009: makes phycobilicyanin out of biliverdin IXalpha
These two parts are thus necessary to make phycobilicyanin, which is the chromophore needed for the proper working of the following part.
- BBa_I15010: makes a chimeric Cph1 light receptor / EnvZ protein
The EnvZ kinase domain phosphorylates OmpR. Now the transcription is initiated by the fourth and last part:
- BBa_R0082: this is a promoter that is activated by phosphorylated OmpR.
Notice that induction of this system could not be accomplished by a light flash, but should be done by a "dark flash". Indeed, red light (660 nm) inhibits phosphorylation of OmpR, and so inhibits the promoter. A dark flash results in OmpR phosphorylation, and so transcription from the OmpR-P dependent promoter. This is however not an ideal system, because keeping the E.coli in 660 nm light environment is not easy.
Notice also that as a result of this system, we need to work with EnvZ deficient E.coli.
Output
Filter
This is what I found (up to now).
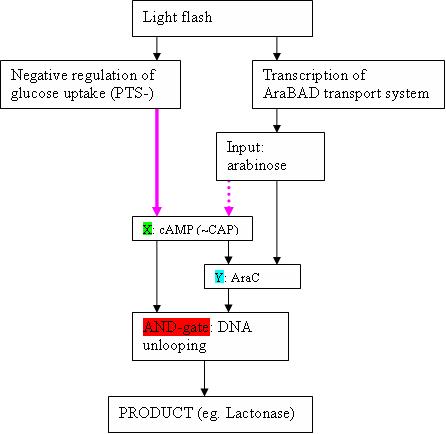
- In the paper of Shen-Orr , the example of the feedforward loop with AND-gate is the well-studied Ara-operon (This can occur when X and Y act in an ‘AND-gate’−like manner to control operon Z, as is the case in the araBAD operon in the arabinose feedforward loop)
- Is dit het enige voorbeeld dat ze aanhalen in shen-orr? misschien is dit systeem nogal moeilijk omvormbaar naar een specifieke filter vermits cAMP nogal een belangrijke rol speelt in de cel – het idee was dan ook om de input van X te veranderen in een induceerbare promoter naar keuze, bijv een OmpR afhankelijke als we het systeen van levskaya met de lichtpuls gebruiken -- IT
- The input signal is of course arabinose (we need AraBAD to transport it efficiently from the medium into the cell)
- What is depicted (see figure) as X, is cAMP (+ CAP)
- Y is AraC
- The AND gate is the “unlooping” of the DNA strand in de Ara operon. This happens only in the presence of both AraC bound to arabinose, and CAP-cAMP
- Product Z are in a WT Ara operon the AraBAD proteïns, but can of course be changed to eg. lactonase. The araBAD genes are located downstream the pBAD promoter, which is activated by the “DNA unlooping”.
- Kan Z ook een tussenproduct zijn dat enerzijds de inverter ingaat richting HSL productie en anderzijds, na het ribolock-key systeem, lactonase productie aanstuurt -- IT
Note that Y is also to be stimulated by arabinose, so the scheme is actually a bit more complex than the easy scheme depicted in the Shen-Orr paper
Some problems:
- I cannot find literature about the dotted purple arrow: is it überhaupt existing? The article of Shen-Orr suggests it does.
- cAMP concentration is depressed by glucose, so maybe the purple arrow means that the concentration of glucose is negligible compared to arabinose. This might however not be good enough.
- What could be done, is making sure that not only AraBAD is made upon (eg) flashing with a light (Austin iGEM 2004), but also making a repressor for the phosphotransferase system of glucose transport. However, then, the pink purple arrow should be the full (non-dotted) pink one.
- I did not (yet) find the BioBricks of the Austin 2004 team for the input signal.
- I believe they can be found here: http://partsregistry.org/Featured_Parts:Light_Sensor -- IT
- Adenylate cyclase inhibition is coupled to the flux of glucose through the glucose PEP:phosphotransferase system . This means that external glucose concentration regulates cAMP production. This is I think a major problem: how do we arrange the system so that it becomes dependent on cytoplasmatic factors?
- Vandaar dat ik hoop dat er nog een ander systeem in E.coli gekend is dat dit netwerkmotief heeft, daar waren we een beetje vanuit gegaan... -- IT
- If cAMP is not degraded fast enough, could cyclic nucleotide phosphodiesterases catalyse this process of degradation?
 "
"